iGTP: a software package for large-scale gene tree parsimony analysis
- PMID: 21092314
- PMCID: PMC3002902
- DOI: 10.1186/1471-2105-11-574
iGTP: a software package for large-scale gene tree parsimony analysis
Abstract
Background: The ever-increasing wealth of genomic sequence information provides an unprecedented opportunity for large-scale phylogenetic analysis. However, species phylogeny inference is obfuscated by incongruence among gene trees due to evolutionary events such as gene duplication and loss, incomplete lineage sorting (deep coalescence), and horizontal gene transfer. Gene tree parsimony (GTP) addresses this issue by seeking a species tree that requires the minimum number of evolutionary events to reconcile a given set of incongruent gene trees. Despite its promise, the use of gene tree parsimony has been limited by the fact that existing software is either not fast enough to tackle large data sets or is restricted in the range of evolutionary events it can handle.
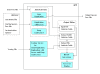
Results: We introduce iGTP, a platform-independent software program that implements state-of-the-art algorithms that greatly speed up species tree inference under the duplication, duplication-loss, and deep coalescence reconciliation costs. iGTP significantly extends and improves the functionality and performance of existing gene tree parsimony software and offers advanced features such as building effective initial trees using stepwise leaf addition and the ability to have unrooted gene trees in the input. Moreover, iGTP provides a user-friendly graphical interface with integrated tree visualization software to facilitate analysis of the results.
Conclusions: iGTP enables, for the first time, gene tree parsimony analyses of thousands of genes from hundreds of taxa using the duplication, duplication-loss, and deep coalescence reconciliation costs, all from within a convenient graphical user interface.
Figures

References
-
- Philip GK, Creevey CJ, McInerney JO. The Opisthokonta and the Ecdysozoa may not be clades: stronger support for the grouping of plant and animal than for animal and fungi and stronger support for the Coelomata than Ecdysozoa. Molecular Biology and Evolution. 2005;22(5):1175–1184. doi: 10.1093/molbev/msi102. - DOI - PubMed

