Stoichiogenomics: the evolutionary ecology of macromolecular elemental composition
- PMID: 21093095
- PMCID: PMC3010507
- DOI: 10.1016/j.tree.2010.10.006
Stoichiogenomics: the evolutionary ecology of macromolecular elemental composition
Abstract
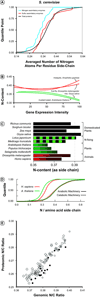
The new field of 'stoichiogenomics' integrates evolution, ecology and bioinformatics to reveal surprising patterns of the differential usage of key elements [e.g. nitrogen (N)] in proteins and nucleic acids. Because the canonical amino acids as well as nucleotides differ in element counts, natural selection owing to limited element supplies might bias monomer usage to reduce element costs. For example, proteins that respond to N limitation in microbes use a lower proportion of N-rich amino acids, whereas proteome- and transcriptome-wide element contents differ significantly for plants as compared with animals, probably because of the differential severity of element limitations. In this review, we show that with these findings, new directions for future investigations are emerging, particularly via the increasing availability of diverse metagenomic and metatranscriptomic data sets.
© 2010 Elsevier Ltd. All rights reserved.
Figures

Similar articles
-
Subcellular stoichiogenomics reveal cell evolution and electrostatic interaction mechanisms in cytoskeleton.BMC Genomics. 2018 Jun 18;19(1):469. doi: 10.1186/s12864-018-4845-0. BMC Genomics. 2018. PMID: 29914356 Free PMC article.
-
Research progress of stoichiogenomics.Yi Chuan. 2017 Feb 20;39(2):89-97. doi: 10.16288/j.yczz.16-343. Yi Chuan. 2017. PMID: 28242596
-
No Evidence That Nitrogen Limitation Influences the Elemental Composition of Isopod Transcriptomes and Proteomes.Mol Biol Evol. 2016 Oct;33(10):2605-20. doi: 10.1093/molbev/msw131. Epub 2016 Jul 8. Mol Biol Evol. 2016. PMID: 27401232
-
Evolutionary Genomics of Plant Gametophytic Selection.Plant Commun. 2020 Oct 24;1(6):100115. doi: 10.1016/j.xplc.2020.100115. eCollection 2020 Nov 9. Plant Commun. 2020. PMID: 33367268 Free PMC article. Review.
-
Urban Evolutionary Ecology and the Potential Benefits of Implementing Genomics.J Hered. 2018 Feb 14;109(2):138-151. doi: 10.1093/jhered/esy001. J Hered. 2018. PMID: 29346589 Review.
Cited by
-
Subcellular stoichiogenomics reveal cell evolution and electrostatic interaction mechanisms in cytoskeleton.BMC Genomics. 2018 Jun 18;19(1):469. doi: 10.1186/s12864-018-4845-0. BMC Genomics. 2018. PMID: 29914356 Free PMC article.
-
Stoichiogenomics reveal oxygen usage bias, key proteins and pathways associated with stomach cancer.Sci Rep. 2019 Aug 5;9(1):11344. doi: 10.1038/s41598-019-47533-6. Sci Rep. 2019. PMID: 31383879 Free PMC article.
-
A Stoichioproteomic Analysis of Samples from the Human Microbiome Project.Front Microbiol. 2017 Jul 18;8:1119. doi: 10.3389/fmicb.2017.01119. eCollection 2017. Front Microbiol. 2017. PMID: 28769875 Free PMC article.
-
Can resource costs of polyploidy provide an advantage to sex?Heredity (Edinb). 2013 Feb;110(2):152-9. doi: 10.1038/hdy.2012.78. Epub 2012 Nov 28. Heredity (Edinb). 2013. PMID: 23188174 Free PMC article.
-
A metabolic prototype for eliminating tryptophan from the genetic code.Sci Rep. 2013;3:1359. doi: 10.1038/srep01359. Sci Rep. 2013. PMID: 23447021 Free PMC article.
References
-
- Eyre-Walker A, Keightley PD. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 2007;8:610–618. - PubMed
-
- Kimura M. The Neutral Theory of Molecular Evolution. Cambridge University Press; 1983.
-
- Wang H-C, et al. On the correlation between genomic G+C content and optimal growth temperature in prokaryotes: Data quality and confounding factors. Biochem. Biophys. Res. Commun. 2006;342:681–684. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

