The structure of the CRISPR-associated protein Csa3 provides insight into the regulation of the CRISPR/Cas system
- PMID: 21093452
- PMCID: PMC4507800
- DOI: 10.1016/j.jmb.2010.11.019
The structure of the CRISPR-associated protein Csa3 provides insight into the regulation of the CRISPR/Cas system
Abstract
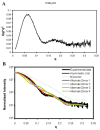
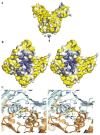
Adaptive immune systems have recently been recognized in prokaryotic organisms where, in response to viral infection, they incorporate short fragments of invader-derived DNA into loci called clustered regularly interspaced short palindromic repeats (CRISPRs). In subsequent infections, the CRISPR loci are transcribed and processed into guide sequences for the neutralization of the invading RNA or DNA. The CRISPR-associated protein machinery (Cas) lies at the heart of this process, yet many of the molecular details of the CRISPR/Cas system remain to be elucidated. Here, we report the first structure of Csa3, a CRISPR-associated protein from Sulfolobus solfataricus (Sso1445), which reveals a dimeric two-domain protein. The N-terminal domain is a unique variation on the dinucleotide binding domain that orchestrates dimer formation. In addition, it utilizes two conserved sequence motifs [Thr-h-Gly-Phe-(Asn/Asp)-Glu-X(4)-Arg and Leu-X(2)-Gly-h-Arg] to construct a 2-fold symmetric pocket on the dimer axis. This pocket is likely to represent a regulatory ligand-binding site. The N-terminal domain is fused to a C-terminal MarR-like winged helix-turn-helix domain that is expected to be involved in DNA recognition. Overall, the unique domain architecture of Csa3 suggests a transcriptional regulator under allosteric control of the N-terminal domain. Alternatively, Csa3 may function in a larger complex, with the conserved cleft participating in protein-protein or protein-nucleic acid interactions. A similar N-terminal domain is also identified in Csx1, a second CRISPR-associated protein family of unknown function.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures
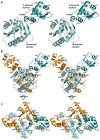
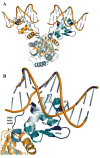

Similar articles
-
SSO1450--a CAS1 protein from Sulfolobus solfataricus P2 with high affinity for RNA and DNA.FEBS Lett. 2009 Jun 18;583(12):1928-32. doi: 10.1016/j.febslet.2009.04.047. Epub 2009 May 8. FEBS Lett. 2009. PMID: 19427858
-
Crystal structure and nucleic acid-binding activity of the CRISPR-associated protein Csx1 of Pyrococcus furiosus.Proteins. 2013 Feb;81(2):261-70. doi: 10.1002/prot.24183. Epub 2012 Oct 16. Proteins. 2013. PMID: 22987782
-
Structure of a dimeric crenarchaeal Cas6 enzyme with an atypical active site for CRISPR RNA processing.Biochem J. 2013 Jun 1;452(2):223-30. doi: 10.1042/BJ20130269. Biochem J. 2013. PMID: 23527601 Free PMC article.
-
Hot and crispy: CRISPR-Cas systems in the hyperthermophile Sulfolobus solfataricus.Biochem Soc Trans. 2013 Dec;41(6):1422-6. doi: 10.1042/BST20130031. Biochem Soc Trans. 2013. PMID: 24256231 Review.
-
The chromosome replication machinery of the archaeon Sulfolobus solfataricus.J Biol Chem. 2006 Jun 2;281(22):15029-32. doi: 10.1074/jbc.R500029200. Epub 2006 Feb 8. J Biol Chem. 2006. PMID: 16467299 Review.
Cited by
-
CRISPR-Associated Factor Csa3b Regulates CRISPR Adaptation and Cmr-Mediated RNA Interference in Sulfolobus islandicus.Front Microbiol. 2020 Aug 26;11:2038. doi: 10.3389/fmicb.2020.02038. eCollection 2020. Front Microbiol. 2020. PMID: 32983033 Free PMC article.
-
Adaptation and modification of three CRISPR loci in two closely related cyanobacteria.RNA Biol. 2013 May;10(5):852-64. doi: 10.4161/rna.24160. Epub 2013 Mar 27. RNA Biol. 2013. PMID: 23535141 Free PMC article.
-
CRISPR-mediated defense mechanisms in the hyperthermophilic archaeal genus Sulfolobus.RNA Biol. 2013 May;10(5):671-8. doi: 10.4161/rna.24154. Epub 2013 Mar 27. RNA Biol. 2013. PMID: 23535277 Free PMC article. Review.
-
The nuts and bolts of the Haloferax CRISPR-Cas system I-B.RNA Biol. 2019 Apr;16(4):469-480. doi: 10.1080/15476286.2018.1460994. Epub 2018 May 21. RNA Biol. 2019. PMID: 29649958 Free PMC article.
-
Microbiology: The case of the mysterious messenger.Nature. 2017 Aug 31;548(7669):527-528. doi: 10.1038/nature23532. Epub 2017 Aug 2. Nature. 2017. PMID: 28783729 No abstract available.
References
-
- van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 2009;34:401–7. - PubMed
-
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–70. - PubMed
-
- Banfield JF, Young M. Microbiology. Variety--the splice of life--in microbial communities. Science. 2009;326:1198–9. - PubMed
-
- Sorek R, Kunin V, Hugenholtz P. CRISPR--a widespread system that provides acquired resistance against phages in bacteria and archaea. Nature reviews Microbiology. 2008;6:181–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

