Ubiquitin ligase switch in plant photomorphogenesis: A hypothesis
- PMID: 21093457
- PMCID: PMC3021735
- DOI: 10.1016/j.jtbi.2010.11.021
Ubiquitin ligase switch in plant photomorphogenesis: A hypothesis
Abstract
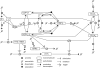
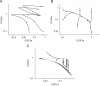
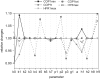
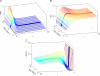
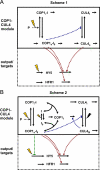
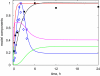
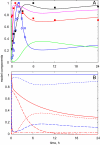
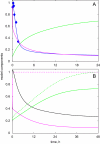
The E3 ubiquitin ligase COP1 (CONSTITUTIVE PHOTOMORPHOGENIC1) plays a key role in the repression of the plant photomorphogenic development in darkness. In the presence of light, COP1 is inactivated by a mechanism which is not completely understood. This leads to accumulation of COP1's target transcription factors, which initiates photomorphogenesis, resulting in dramatic changes of the seedling's physiology. Here we use a mathematical model to explore the possible mechanism of COP1 modulation upon dark/light transition in Arabidopsis thaliana based upon data for two COP1 target proteins: HY5 and HFR1, which play critical roles in photomorphogenesis. The main reactions in our model are the inactivation of COP1 by a proposed photoreceptor-related inhibitor I and interactions between COP1 and a CUL4 (CULLIN4)-based ligase. For building and verification of the model, we used the available published and our new data on the kinetics of HY5 and HFR1 together with the data on COP1 abundance. HY5 has been shown to accumulate at a slower rate than HFR1. To describe the observed differences in the timecourses of the "slow" target HY5 and the "fast" target HFR1, we hypothesize a switch between the activities of COP1 and CUL4 ligases upon dark/light transition, with COP1 being active mostly in darkness and CUL4 in light. The model predicts a bi-phasic kinetics of COP1 activity upon the exposure of plants to light, with its restoration after the initial decline and the following slow depletion of the total COP1 content. CUL4 activity is predicted to increase in the presence of light. We propose that the ubiquitin ligase switch is important for the complex regulation of multiple transcription factors during plants development. In addition, this provides a new mechanism for sensing the duration of light period, which is important for seasonal changes in plant development.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures

Similar articles
-
Arabidopsis CULLIN4 Forms an E3 Ubiquitin Ligase with RBX1 and the CDD Complex in Mediating Light Control of Development.Plant Cell. 2006 Aug;18(8):1991-2004. doi: 10.1105/tpc.106.043224. Epub 2006 Jul 14. Plant Cell. 2006. PMID: 16844902 Free PMC article.
-
The E3 ubiquitin ligases RING DOMAIN OF UNKNOWN FUNCTION 1117 1 (RDUF1) and RDUF2 control seedling photomorphogenesis in Arabidopsis.New Phytol. 2025 Jul;247(2):684-705. doi: 10.1111/nph.70169. Epub 2025 May 21. New Phytol. 2025. PMID: 40396444
-
Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis.Plant Cell. 2005 Mar;17(3):804-21. doi: 10.1105/tpc.104.030205. Epub 2005 Feb 10. Plant Cell. 2005. PMID: 15705947 Free PMC article.
-
The photomorphogenic repressors COP1 and DET1: 20 years later.Trends Plant Sci. 2012 Oct;17(10):584-93. doi: 10.1016/j.tplants.2012.05.004. Epub 2012 Jun 15. Trends Plant Sci. 2012. PMID: 22705257 Review.
-
The molecular basis of CONSTITUTIVE PHOTOMORPHOGENIC1 action during photomorphogenesis.J Exp Bot. 2025 Feb 7;76(3):664-676. doi: 10.1093/jxb/erae181. J Exp Bot. 2025. PMID: 38683181 Review.
Cited by
-
Photomorphogenesis.Arabidopsis Book. 2012;10:e0147. doi: 10.1199/tab.0147. Epub 2012 Jan 31. Arabidopsis Book. 2012. PMID: 22582028 Free PMC article.
-
Abundant clock proteins point to missing molecular regulation in the plant circadian clock.Mol Syst Biol. 2025 Apr;21(4):361-389. doi: 10.1038/s44320-025-00086-5. Epub 2025 Feb 20. Mol Syst Biol. 2025. PMID: 39979593 Free PMC article.
-
ClPIF3-ClHY5 Module Regulates ClPSY1 to Promote Watermelon Fruit Lycopene Accumulation Earlier under Supplementary Red Lighting.Int J Mol Sci. 2022 Apr 8;23(8):4145. doi: 10.3390/ijms23084145. Int J Mol Sci. 2022. PMID: 35456963 Free PMC article.
-
The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops.Mol Syst Biol. 2012 Mar 6;8:574. doi: 10.1038/msb.2012.6. Mol Syst Biol. 2012. PMID: 22395476 Free PMC article.
-
Modelling the widespread effects of TOC1 signalling on the plant circadian clock and its outputs.BMC Syst Biol. 2013 Mar 19;7:23. doi: 10.1186/1752-0509-7-23. BMC Syst Biol. 2013. PMID: 23506153 Free PMC article.
References
-
- Ang L.H., Chattopadhyay S., Wei N., Oyama T., Okada K., Batschauer A., Deng X.W. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell. 1998;1:213–222. - PubMed
-
- Chamovitz D.A., Wei N., Osterlund M.T., von Arnim A.G., Staub J.M., Matsui M., Deng X.W. The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell. 1996;86:115–121. - PubMed
-
- Chen H., Huang X., Gusmaroli G., Terzaghi W., Lau O.S., Yanagawa Y., Zhang Y., Li J., Lee J.H., Zhu D., Deng X.W. Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell. 2010;22:108–123. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

