Synthetic lethality: general principles, utility and detection using genetic screens in human cells
- PMID: 21094158
- PMCID: PMC3018572
- DOI: 10.1016/j.febslet.2010.11.024
Synthetic lethality: general principles, utility and detection using genetic screens in human cells
Abstract
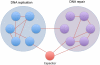
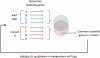
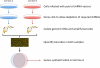
Synthetic lethality occurs when the simultaneous perturbation of two genes results in cellular or organismal death. Synthetic lethality also occurs between genes and small molecules, and can be used to elucidate the mechanism of action of drugs. This area has recently attracted attention because of the prospect of a new generation of anti-cancer drugs. Based on studies ranging from yeast to human cells, this review provides an overview of the general principles that underlie synthetic lethality and relates them to its utility for identifying gene function, drug action and cancer therapy. It also identifies the latest strategies for the large-scale mapping of synthetic lethalities in human cells which bring us closer to the generation of comprehensive human genetic interaction maps.
Copyright © 2010 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures


References
-
- Bridges C.B. The origin of variation. Amer Nat. 1922;56:51–63.
-
- Boone C., Bussey H., Andrews B.J. Exploring genetic interactions and networks with yeast. Nat Rev Genet. 2007;8:437–449. - PubMed
-
- Hartman J.L.T., Garvik B., Hartwell L. Principles for the buffering of genetic variation. Science. 2001;291:1001–1004. - PubMed
-
- Kaelin W.G., Jr. The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–698. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

