Comparative analysis of cleavable azobenzene-based affinity tags for bioorthogonal chemical proteomics
- PMID: 21095571
- PMCID: PMC3103785
- DOI: 10.1016/j.chembiol.2010.09.012
Comparative analysis of cleavable azobenzene-based affinity tags for bioorthogonal chemical proteomics
Abstract
The advances in bioorthogonal ligation methods have provided new opportunities for proteomic analysis of newly synthesized proteins, posttranslational modifications, and specific enzyme families using azide/alkyne-functionalized chemical reporters and activity-based probes. Efficient enrichment and elution of azide/alkyne-labeled proteins with selectively cleavable affinity tags are essential for protein identification and quantification applications. Here, we report the synthesis and comparative analysis of Na₂S₂O₄-cleavable azobenzene-based affinity tags for bioorthogonal chemical proteomics. We demonstrated that ortho-hydroxyl substituent is required for efficient azobenzene-bond cleavage and show that these cleavable affinity tags can be used to identify newly synthesized proteins in bacteria targeted by amino acid chemical reporters as well as their sites of modification on endogenously expressed proteins. The azobenzene-based affinity tags are compatible with in-gel, in-solution, and on-bead enrichment strategies and should afford useful tools for diverse bioorthogonal proteomic applications.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures




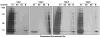


Comment in
-
Reeling in the catch: advancing cleavable linkers for proteomics.Chem Biol. 2010 Nov 24;17(11):1166-8. doi: 10.1016/j.chembiol.2010.11.001. Chem Biol. 2010. PMID: 21095564
References
-
- Bahulayan DJ, Lalithambika LM. Modified clays as efficient acid-base catalyst systems for diazotization and diazocoupling reactions. Synth Comm. 2003;33:863–869.
-
- Best MD. Click chemistry and bioorthogonal reactions: unprecedented selectivity in the labeling of biological molecules. Biochemistry. 2009;48:6571–6584. - PubMed
-
- Böttcher T, Pitscheider M, Sieber SA. Natural products and their biological targets: proteomic and metabolomic labeling strategies. Angew Chem Int Ed. 2010;49:2680–2698. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

