Ligand-induced conformational capture of a synthetic tetracycline riboswitch revealed by pulse EPR
- PMID: 21097555
- PMCID: PMC3004059
- DOI: 10.1261/rna.2222811
Ligand-induced conformational capture of a synthetic tetracycline riboswitch revealed by pulse EPR
Abstract
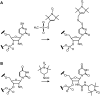
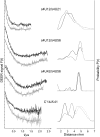
RNA aptamers are in vitro-selected binding domains that recognize their respective ligand with high affinity and specificity. They are characterized by complex three-dimensional conformations providing preformed binding pockets that undergo conformational changes upon ligand binding. Small molecule-binding aptamers have been exploited as synthetic riboswitches for conditional gene expression in various organisms. In the present study, double electron-electron resonance (DEER) spectroscopy combined with site-directed spin labeling was used to elucidate the conformational transition of a tetracycline aptamer upon ligand binding. Different sites were selected for post-synthetic introduction of either the (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl) methanethiosulfonate by reaction with a 4-thiouridine modified RNA or of 4-isocyanato-2,6-tetramethylpiperidyl-N-oxid spin label by reaction with 2'-aminouridine modified RNA. The results of the DEER experiments indicate the presence of a thermodynamic equilibrium between two aptamer conformations in the free state and capture of one conformation upon tetracycline binding.
Figures



References
-
- Berens C, Hillen W 2003. Gene regulation by tetracyclines. Eur J Biochem 270: 3109–3121 - PubMed
-
- Bordignon E, Steinhoff HJ 2007. Membrane protein structure and dynamics studied by site-directed spin labeling ESR. In ESR spectroscopy in membrane biophysics (ed. Hemminga MA, Berliner LJ), pp. 129–164 Springer Science and Business Media, New York
-
- Edwards TE, Okonogi TM, Robinson BH, Sigurdsson ST 2001. Site-specific incorporation of nitroxide spin-labels into internal sites of the TAR RNA; structure-dependent dynamics of RNA by EPR spectroscopy. J Am Chem Soc 123: 1527–1528 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
