L-malate production by metabolically engineered Escherichia coli
- PMID: 21097588
- PMCID: PMC3020529
- DOI: 10.1128/AEM.01971-10
L-malate production by metabolically engineered Escherichia coli
Abstract
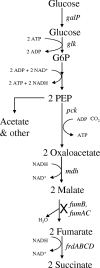
Escherichia coli strains (KJ060 and KJ073) that were previously developed for succinate production have now been modified for malate production. Many unexpected changes were observed during this investigation. The initial strategy of deleting fumarase isoenzymes was ineffective, and succinate continued to accumulate. Surprisingly, a mutation in fumarate reductase alone was sufficient to redirect carbon flow into malate even in the presence of fumarase. Further deletions were needed to inactivate malic enzymes (typically gluconeogenic) and prevent conversion to pyruvate. However, deletion of these genes (sfcA and maeB) resulted in the unexpected accumulation of D-lactate despite the prior deletion of mgsA and ldhA and the absence of apparent lactate dehydrogenase activity. Although the metabolic source of this D-lactate was not identified, lactate accumulation was increased by supplementation with pyruvate and decreased by the deletion of either pyruvate kinase gene (pykA or pykF) to reduce the supply of pyruvate. Many of the gene deletions adversely affected growth and cell yield in minimal medium under anaerobic conditions, and volumetric rates of malate production remained low. The final strain (XZ658) produced 163 mM malate, with a yield of 1.0 mol (mol glucose(-1)), half of the theoretical maximum. Using a two-stage process (aerobic cell growth and anaerobic malate production), this engineered strain produced 253 mM malate (34 g liter(-1)) within 72 h, with a higher yield (1.42 mol mol(-1)) and productivity (0.47 g liter(-1) h(-1)). This malate yield and productivity are equal to or better than those of other known biocatalysts.
Figures
References
-
- Atsumi, S., T. Hanai, and J. C. Liao. 2008. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86-89. - PubMed
-
- Battat, E., Y. Peleg, A. Bercovitz, J. S. Rokem, and I. Goldberg. 1991. Optimization of l-malic acid production by Aspergillus flavus in a stirred fermentor. Biotechnol. Bioeng. 37:1108-1116. - PubMed
-
- Bressler, E., O. Pines, I. Goldberg, and S. Braun. 2002. Conversion of fumaric acid to l-malic by sol-gel immobilized Saccharomyces cerevisiae in a supported liquid membrane bioreactor. Biotechnol. Prog. 18:445-450. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

