Metatranscriptome analysis for insight into whole-ecosystem gene expression during spontaneous wheat and spelt sourdough fermentations
- PMID: 21097589
- PMCID: PMC3020550
- DOI: 10.1128/AEM.02028-10
Metatranscriptome analysis for insight into whole-ecosystem gene expression during spontaneous wheat and spelt sourdough fermentations
Abstract
Lactic acid bacteria (LAB) are of industrial importance in the production of fermented foods, including sourdough-derived products. Despite their limited metabolic capacity, LAB contribute considerably to important characteristics of fermented foods, such as extended shelf-life, microbial safety, improved texture, and enhanced organoleptic properties. Triggered by the considerable amount of LAB genomic information that became available during the last decade, transcriptome and, by extension, metatranscriptome studies have become one of the most appropriate research approaches to study whole-ecosystem gene expression in more detail. In this study, microarray analyses were performed using RNA sampled during four 10-day spontaneous sourdough fermentations carried out in the laboratory with an in-house-developed LAB functional gene microarray. For data analysis, a new algorithm was developed to calculate a net expression profile for each of the represented genes, allowing use of the microarray analysis beyond the species level. In addition, metabolite target analyses were performed on the sourdough samples to relate gene expression with metabolite production. The results revealed the activation of different key metabolic pathways, the ability to use carbohydrates other than glucose (e.g., starch and maltose), and the conversion of amino acids as a contribution to redox equilibrium and flavor compound generation in LAB during sourdough fermentation.
Figures
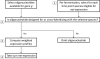

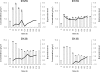
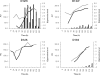
References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

