Isolation and characterization of P1 adhesin, a leg protein of the gliding bacterium Mycoplasma pneumoniae
- PMID: 21097617
- PMCID: PMC3021217
- DOI: 10.1128/JB.00796-10
Isolation and characterization of P1 adhesin, a leg protein of the gliding bacterium Mycoplasma pneumoniae
Abstract
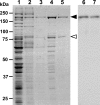
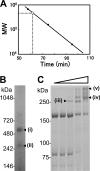
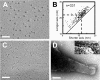
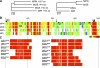
Mycoplasma pneumoniae, a pathogen causing human pneumonia, binds to solid surfaces at its membrane protrusion and glides by a unique mechanism. In this study, P1 adhesin, which functions as a "leg" in gliding, was isolated from mycoplasma culture and characterized. Using gel filtration, blue-native polyacrylamide gel electrophoresis (BN-PAGE), and chemical cross-linking, the isolated P1 adhesin was shown to form a complex with an accessory protein named P90. The complex included two molecules each of P1 adhesin and P90 (protein B), had a molecular mass of about 480 kDa, and was observed by electron microscopy to form 20-nm-diameter spheres. Partial digestion of isolated P1 adhesin by trypsin showed that the P1 adhesin molecule can be divided into three domains, consistent with the results from trypsin treatment of the cell surface. Sequence analysis of P1 adhesin and its orthologs showed that domain I is well conserved and that a transmembrane segment exists near the link between domains II and III.
Figures


References
-
- Aluotto, B. B., R. G. Wittler, C. O. Williams, and J. E. Faber. 1970. Standardized bacteriologic techniques for the characterization of mycoplasma species. Int. J. Syst. Bacteriol. 20:35-58.
-
- Arata, T. 1998. Electron microscopic observation of monomeric actin attached to a myosin head. J. Struct. Biol. 123:8-16. - PubMed
-
- Catrein, I., R. Herrmann, A. Bosserhoff, and T. Ruppert. 2005. Experimental proof for a signal peptidase I like activity in Mycoplasma pneumoniae, but absence of a gene encoding a conserved bacterial type I SPase. FEBS J. 272:2892-2900. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

