Chemotaxis to the quorum-sensing signal AI-2 requires the Tsr chemoreceptor and the periplasmic LsrB AI-2-binding protein
- PMID: 21097621
- PMCID: PMC3021223
- DOI: 10.1128/JB.01196-10
Chemotaxis to the quorum-sensing signal AI-2 requires the Tsr chemoreceptor and the periplasmic LsrB AI-2-binding protein
Abstract
AI-2 is an autoinducer made by many bacteria. LsrB binds AI-2 in the periplasm, and Tsr is the l-serine chemoreceptor. We show that AI-2 strongly attracts Escherichia coli. Both LsrB and Tsr are necessary for sensing AI-2, but AI-2 uptake is not, suggesting that LsrB and Tsr interact directly in the periplasm.
Figures
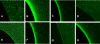

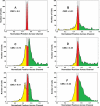
References
-
- Adler, J. 1973. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J. Gen. Microbiol. 74:77-91. - PubMed
-
- Aksamit, R. R., and D. E. Koshland, Jr. 1974. Identification of the ribose binding protein as the receptor for ribose chemotaxis in Salmonella typhimurium. Biochemistry 13:4473-4478. - PubMed
-
- Bansal, T., P. Jesudhasan, S. Pillai, T. K. Wood, and A. Jayaraman. 2008. Temporal regulation of enterohemorrhagic Escherichia coli virulence mediated by autoinducer-2. Appl. Microbiol. Biotechnol. 78:811-819. - PubMed
-
- Bassler, B. L. 2002. Small talk: cell-to-cell communication in bacteria. Cell 109:421-424. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

