Altered regulation of the OmpF porin by Fis in Escherichia coli during an evolution experiment and between B and K-12 strains
- PMID: 21097626
- PMCID: PMC3019833
- DOI: 10.1128/JB.01341-10
Altered regulation of the OmpF porin by Fis in Escherichia coli during an evolution experiment and between B and K-12 strains
Abstract
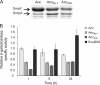
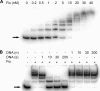
The phenotypic plasticity of global regulatory networks provides bacteria with rapid acclimation to a wide range of environmental conditions, while genetic changes in those networks provide additional flexibility as bacteria evolve across long time scales. We previously identified mutations in the global regulator-encoding gene fis that enhanced organismal fitness during a long-term evolution experiment with Escherichia coli. To gain insight into the effects of these mutations, we produced two-dimensional protein gels with strains carrying different fis alleles, including a beneficial evolved allele and one with an in-frame deletion. We found that Fis controls the expression of the major porin-encoding gene ompF in the E. coli B-derived ancestral strain used in the evolution experiment, a relationship that has not been described before. We further showed that this regulatory connection evolved over two different time scales, perhaps explaining why it was not observed before. On the longer time scale, we showed that this regulation of ompF by Fis is absent from the more widely studied K-12 strain and thus is specific to the B strain. On a shorter time scale, this regulatory linkage was lost during 20,000 generations of experimental evolution of the B strain. Finally, we mapped the Fis binding sites in the ompF regulatory region, and we present a hypothetical model of ompF expression that includes its other known regulators.
Figures




Similar articles
-
Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli.Nucleic Acids Res. 2011 Mar;39(6):2073-91. doi: 10.1093/nar/gkq934. Epub 2010 Nov 21. Nucleic Acids Res. 2011. PMID: 21097887 Free PMC article.
-
Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts.Genome Res. 2008 Jun;18(6):900-10. doi: 10.1101/gr.070276.107. Epub 2008 Mar 13. Genome Res. 2008. PMID: 18340041 Free PMC article.
-
The role of FIS protein in the physiological control of the expression of the Escherichia coli meta-hpa operon.Microbiology (Reading). 2008 Jul;154(Pt 7):2151-2160. doi: 10.1099/mic.0.2007/015578-0. Microbiology (Reading). 2008. PMID: 18599842
-
IHF and Fis as Escherichia coli Cell Cycle Regulators: Activation of the Replication Origin oriC and the Regulatory Cycle of the DnaA Initiator.Int J Mol Sci. 2023 Jul 18;24(14):11572. doi: 10.3390/ijms241411572. Int J Mol Sci. 2023. PMID: 37511331 Free PMC article. Review.
-
Molecular basis of voltage gating of OmpF porin.Biochem Cell Biol. 2002;80(5):517-23. doi: 10.1139/o02-145. Biochem Cell Biol. 2002. PMID: 12440693 Review.
Cited by
-
Gene knockout studies of Dps protein reveals a novel role for DNA-binding protein in maintaining outer membrane permeability.World J Microbiol Biotechnol. 2025 Feb 13;41(2):70. doi: 10.1007/s11274-025-04269-y. World J Microbiol Biotechnol. 2025. PMID: 39939516 Free PMC article.
-
Repeatability and contingency in the evolution of a key innovation in phage lambda.Science. 2012 Jan 27;335(6067):428-32. doi: 10.1126/science.1214449. Science. 2012. PMID: 22282803 Free PMC article.
-
Synonymous Mutations in rpsT Lead to Ribosomal Assembly Defects That Can Be Compensated by Mutations in fis and rpoA.Front Microbiol. 2020 Mar 6;11:340. doi: 10.3389/fmicb.2020.00340. eCollection 2020. Front Microbiol. 2020. PMID: 32210939 Free PMC article.
-
Proteome partitioning constraints in long-term laboratory evolution.Nat Commun. 2024 May 14;15(1):4087. doi: 10.1038/s41467-024-48447-2. Nat Commun. 2024. PMID: 38744842 Free PMC article.
-
AmpC hyperproduction in a Cedecea davisae implant-associated bone infection during treatment: a case report and therapeutic implications.BMC Infect Dis. 2022 Jan 6;22(1):33. doi: 10.1186/s12879-021-07000-y. BMC Infect Dis. 2022. PMID: 34991516 Free PMC article.
References
-
- Barrick, J. E., et al. 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461:1243-1247. - PubMed
-
- Bergstrom, L. C., L. Qin, S. L. Harlocker, L. A. Egger, and M. Inouye. 1998. Hierarchical and cooperative binding of OmpR to a fusion construct containing the ompC and ompF upstream regulatory sequences of Escherichia coli. Genes Cells 3:777-788. - PubMed
-
- Bocharov, E. V., et al. 2004. From structure and dynamics of protein L7/L12 to molecular switching in ribosome. J. Biol. Chem. 279:17697-17706. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

