Tertiary contacts control switching of the SAM-I riboswitch
- PMID: 21097777
- PMCID: PMC3064774
- DOI: 10.1093/nar/gkq1096
Tertiary contacts control switching of the SAM-I riboswitch
Abstract
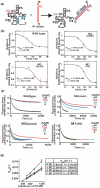
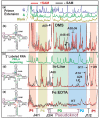
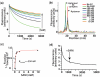
Riboswitches are non-coding RNAs that control gene expression by sensing small molecules through changes in secondary structure. While secondary structure and ligand interactions are thought to control switching, the exact mechanism of control is unknown. Using a novel two-piece assay that competes the anti-terminator against the aptamer, we directly monitor the process of switching. We find that the stabilization of key tertiary contacts controls both aptamer domain collapse and the switching of the SAM-I riboswitch from the aptamer to the expression platform conformation. Our experiments demonstrate that SAM binding induces structural alterations that indirectly stabilize the aptamer domain, preventing switching toward the expression platform conformer. These results, combined with a variety of structural probing experiments performed in this study, show that the collapse and stabilization of the aptamer domain are cooperative, relying on the sum of key tertiary contacts and the bimodal stability of the kink-turn motif for function. Here, ligand binding serves to shift the equilibrium of aptamer domain structures from a more open toward a more stable collapsed form by stabilizing tertiary interactions. Our data show that the thermodynamic landscape for riboswitch operation is finely balanced to allow large conformational rearrangements to be controlled by small molecule interactions.
Figures




Similar articles
-
The expression platform and the aptamer: cooperativity between Mg2+ and ligand in the SAM-I riboswitch.Nucleic Acids Res. 2013 Feb 1;41(3):1922-35. doi: 10.1093/nar/gks978. Epub 2012 Dec 20. Nucleic Acids Res. 2013. PMID: 23258703 Free PMC article.
-
Basis for ligand discrimination between ON and OFF state riboswitch conformations: the case of the SAM-I riboswitch.RNA. 2012 Jun;18(6):1230-43. doi: 10.1261/rna.032177.111. Epub 2012 Apr 27. RNA. 2012. PMID: 22543867 Free PMC article.
-
RNA Ensembles from Solvent Accessibility Data: Application to the SAM-I Riboswitch Aptamer Domain.J Phys Chem B. 2021 Apr 15;125(14):3486-3493. doi: 10.1021/acs.jpcb.0c11503. Epub 2021 Apr 5. J Phys Chem B. 2021. PMID: 33818089
-
Structural studies of the purine and SAM binding riboswitches.Cold Spring Harb Symp Quant Biol. 2006;71:259-68. doi: 10.1101/sqb.2006.71.015. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381305 Review.
-
Structure-based insights into recognition and regulation of SAM-sensing riboswitches.Sci China Life Sci. 2023 Jan;66(1):31-50. doi: 10.1007/s11427-022-2188-7. Epub 2022 Nov 29. Sci China Life Sci. 2023. PMID: 36459353 Review.
Cited by
-
Magnesium fluctuations modulate RNA dynamics in the SAM-I riboswitch.J Am Chem Soc. 2012 Jul 25;134(29):12043-53. doi: 10.1021/ja301454u. Epub 2012 Jul 16. J Am Chem Soc. 2012. PMID: 22612276 Free PMC article.
-
Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution.Nat Commun. 2020 Jan 9;11(1):148. doi: 10.1038/s41467-019-13942-4. Nat Commun. 2020. PMID: 31919376 Free PMC article.
-
Exploring the Energy Landscape of Riboswitches Using Collective Variables Based on Tertiary Contacts.J Mol Biol. 2022 Sep 30;434(18):167788. doi: 10.1016/j.jmb.2022.167788. Epub 2022 Aug 11. J Mol Biol. 2022. PMID: 35963460 Free PMC article.
-
Magnesium controls aptamer-expression platform switching in the SAM-I riboswitch.Nucleic Acids Res. 2019 Apr 8;47(6):3158-3170. doi: 10.1093/nar/gky1311. Nucleic Acids Res. 2019. PMID: 30605518 Free PMC article.
-
Dynamics of riboswitches: Molecular simulations.Biochim Biophys Acta. 2014 Oct;1839(10):1046-1050. doi: 10.1016/j.bbagrm.2014.06.010. Epub 2014 Jun 19. Biochim Biophys Acta. 2014. PMID: 24953187 Free PMC article. Review.
References
-
- Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. - PubMed
-
- Grundy FJ, Henkin TM. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 1998;30:737–749. - PubMed
-
- Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 2003;10:701–707. - PubMed

