Extending CATH: increasing coverage of the protein structure universe and linking structure with function
- PMID: 21097779
- PMCID: PMC3013636
- DOI: 10.1093/nar/gkq1001
Extending CATH: increasing coverage of the protein structure universe and linking structure with function
Abstract
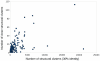
CATH version 3.3 (class, architecture, topology, homology) contains 128,688 domains, 2386 homologous superfamilies and 1233 fold groups, and reflects a major focus on classifying structural genomics (SG) structures and transmembrane proteins, both of which are likely to add structural novelty to the database and therefore increase the coverage of protein fold space within CATH. For CATH version 3.4 we have significantly improved the presentation of sequence information and associated functional information for CATH superfamilies. The CATH superfamily pages now reflect both the functional and structural diversity within the superfamily and include structural alignments of close and distant relatives within the superfamily, annotated with functional information and details of conserved residues. A significantly more efficient search function for CATH has been established by implementing the search server Solr (http://lucene.apache.org/solr/). The CATH v3.4 webpages have been built using the Catalyst web framework.
Figures

References
-
- Orengo CA, Michie AD, Jones S, Jones DT, Swindells MB, Thornton JM. CATH – a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1109. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

