Cooperation of RAD51 and RAD54 in regression of a model replication fork
- PMID: 21097884
- PMCID: PMC3064783
- DOI: 10.1093/nar/gkq1139
Cooperation of RAD51 and RAD54 in regression of a model replication fork
Abstract
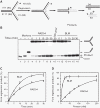
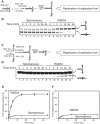
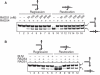
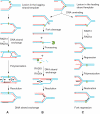
DNA lesions cause stalling of DNA replication forks, which can be lethal for the cell. Homologous recombination (HR) plays an important role in DNA lesion bypass. It is thought that Rad51, a key protein of HR, contributes to the DNA lesion bypass through its DNA strand invasion activity. Here, using model stalled replication forks we found that RAD51 and RAD54 by acting together can promote DNA lesion bypass in vitro through the 'template-strand switch' mechanism. This mechanism involves replication fork regression into a Holliday junction ('chicken foot structure'), DNA synthesis using the nascent lagging DNA strand as a template and fork restoration. Our results demonstrate that RAD54 can catalyze both regression and restoration of model replication forks through its branch migration activity, but shows strong bias toward fork restoration. We find that RAD51 modulates this reaction; by inhibiting fork restoration and stimulating fork regression it promotes accumulation of the chicken foot structure, which we show is essential for DNA lesion bypass by DNA polymerase in vitro. These results indicate that RAD51 in cooperation with RAD54 may have a new role in DNA lesion bypass that is distinct from DNA strand invasion.
Figures



References
-
- Rothstein R, Michel B, Gangloff S. Replication fork pausing and recombination or ‘gimme a break’. Genes Dev. 2000;14:1–10. - PubMed
-
- Budzowska M, Kanaar R. Mechanisms of dealing with DNA damage-induced replication problems. Cell Biochem. Biophys. 2009;53:17–31. - PubMed
-
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. - PubMed
-
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

