Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli
- PMID: 21097887
- PMCID: PMC3064808
- DOI: 10.1093/nar/gkq934
Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli
Abstract
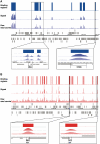
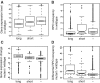
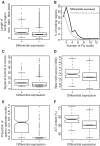
Nucleoid-associated proteins (NAPs) are global regulators of gene expression in Escherichia coli, which affect DNA conformation by bending, wrapping and bridging the DNA. Two of these--H-NS and Fis--bind to specific DNA sequences and structures. Because of their importance to global gene expression, the binding of these NAPs to the DNA was previously investigated on a genome-wide scale using ChIP-chip. However, variation in their binding profiles across the growth phase and the genome-scale nature of their impact on gene expression remain poorly understood. Here, we present a genome-scale investigation of H-NS and Fis binding to the E. coli chromosome using chromatin immunoprecipitation combined with high-throughput sequencing (ChIP-seq). By performing our experiments under multiple time-points during growth in rich media, we show that the binding regions of the two proteins are mutually exclusive under our experimental conditions. H-NS binds to significantly longer tracts of DNA than Fis, consistent with the linear spread of H-NS binding from high- to surrounding lower-affinity sites; the length of binding regions is associated with the degree of transcriptional repression imposed by H-NS. For Fis, a majority of binding events do not lead to differential expression of the proximal gene; however, it has a significant indirect effect on gene expression partly through its effects on the expression of other transcription factors. We propose that direct transcriptional regulation by Fis is associated with the interaction of tandem arrays of Fis molecules to the DNA and possible DNA bending, particularly at operon-upstream regions. Our study serves as a proof-of-principle for the use of ChIP-seq for global DNA-binding proteins in bacteria, which should become significantly more economical and feasible with the development of multiplexing techniques.
Figures





Similar articles
-
Reconstruction of transcriptional regulatory networks of Fis and H-NS in Escherichia coli from genome-wide data analysis.Genomics. 2020 Mar;112(2):1264-1272. doi: 10.1016/j.ygeno.2019.07.013. Epub 2019 Jul 26. Genomics. 2020. PMID: 31356968
-
Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts.Genome Res. 2008 Jun;18(6):900-10. doi: 10.1101/gr.070276.107. Epub 2008 Mar 13. Genome Res. 2008. PMID: 18340041 Free PMC article.
-
Chromosome organization by a nucleoid-associated protein in live bacteria.Science. 2011 Sep 9;333(6048):1445-9. doi: 10.1126/science.1204697. Science. 2011. PMID: 21903814 Free PMC article.
-
Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression.Curr Opin Microbiol. 2010 Dec;13(6):773-80. doi: 10.1016/j.mib.2010.09.013. Epub 2010 Oct 13. Curr Opin Microbiol. 2010. PMID: 20951079 Review.
-
Structure and function of bacterial H-NS protein.Biochem Soc Trans. 2016 Dec 15;44(6):1561-1569. doi: 10.1042/BST20160190. Biochem Soc Trans. 2016. PMID: 27913665 Review.
Cited by
-
Promoters of Escherichia coli versus promoter islands: function and structure comparison.PLoS One. 2013 May 22;8(5):e62601. doi: 10.1371/journal.pone.0062601. Print 2013. PLoS One. 2013. PMID: 23717391 Free PMC article.
-
Site-specific DNA Inversion by Serine Recombinases.Microbiol Spectr. 2015 Feb 19;3(3):1-36. doi: 10.1128/microbiolspec.MDNA3-0047-2014. Microbiol Spectr. 2015. PMID: 25844275 Free PMC article.
-
An Interplay among FIS, H-NS, and Guanosine Tetraphosphate Modulates Transcription of the Escherichia coli cspA Gene under Physiological Growth Conditions.Front Mol Biosci. 2016 May 24;3:19. doi: 10.3389/fmolb.2016.00019. eCollection 2016. Front Mol Biosci. 2016. PMID: 27252944 Free PMC article.
-
H-NS is a novel transcriptional modulator of the ribonucleotide reductase genes in Escherichia coli.J Bacteriol. 2013 Sep;195(18):4255-63. doi: 10.1128/JB.00490-13. Epub 2013 Jul 19. J Bacteriol. 2013. PMID: 23873909 Free PMC article.
-
Spatiotemporal Coupling of DNA Supercoiling and Genomic Sequence Organization-A Timing Chain for the Bacterial Growth Cycle?Biomolecules. 2022 Jun 15;12(6):831. doi: 10.3390/biom12060831. Biomolecules. 2022. PMID: 35740956 Free PMC article. Review.
References
-
- Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004;2:57–65. - PubMed
-
- Martínez-Antonio A, Collado-Vides J. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 2003;6:482–489. - PubMed
-
- Dillon SC, Dorman CJ. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010;8:185–195. - PubMed
-
- Dorman CJ, Kane KA. DNA bridging and antibridging: a role for bacterial nucleoid-associated proteins in regulating the expression of laterally acquired genes. FEMS Microbiol. Rev. 2009;33:587–592. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

