NCBI GEO: archive for functional genomics data sets--10 years on
- PMID: 21097893
- PMCID: PMC3013736
- DOI: 10.1093/nar/gkq1184
NCBI GEO: archive for functional genomics data sets--10 years on
Abstract
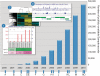
A decade ago, the Gene Expression Omnibus (GEO) database was established at the National Center for Biotechnology Information (NCBI). The original objective of GEO was to serve as a public repository for high-throughput gene expression data generated mostly by microarray technology. However, the research community quickly applied microarrays to non-gene-expression studies, including examination of genome copy number variation and genome-wide profiling of DNA-binding proteins. Because the GEO database was designed with a flexible structure, it was possible to quickly adapt the repository to store these data types. More recently, as the microarray community switches to next-generation sequencing technologies, GEO has again adapted to host these data sets. Today, GEO stores over 20,000 microarray- and sequence-based functional genomics studies, and continues to handle the majority of direct high-throughput data submissions from the research community. Multiple mechanisms are provided to help users effectively search, browse, download and visualize the data at the level of individual genes or entire studies. This paper describes recent database enhancements, including new search and data representation tools, as well as a brief review of how the community uses GEO data. GEO is freely accessible at http://www.ncbi.nlm.nih.gov/geo/.
Figures
References
-
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 2001;29:365–371. - PubMed
-
- Microarray standards at last. Nature. 2002;419:323. Available at http://www.nature.com/nature/journal/v419/n6905/full/419323a.html. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

