Phenotypic and genotypic characterization of influenza virus mutants selected with the sialidase fusion protein DAS181
- PMID: 21097900
- PMCID: PMC3001847
- DOI: 10.1093/jac/dkq387
Phenotypic and genotypic characterization of influenza virus mutants selected with the sialidase fusion protein DAS181
Abstract
Background: influenza viruses (IFVs) frequently achieve resistance to antiviral drugs, necessitating the development of compounds with novel mechanisms of action. DAS181 (Fludase), a sialidase fusion protein, may have a reduced potential for generating drug resistance due to its novel host-targeting mechanism of action.
Methods: IFV strains B/Maryland/1/59 and A/Victoria/3/75 (H3N2) were subjected to >30 passages under increasing selective pressure with DAS181. The DAS181-selected IFV isolates were characterized in vitro and in mice.
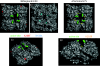
Results: despite extensive passaging, DAS181-selected viruses exhibited a very low level of resistance to DAS181, which ranged between 3- and 18-fold increase in EC(50). DAS181-selected viruses displayed an attenuated phenotype in vitro, as exhibited by slower growth, smaller plaque size and increased particle to pfu ratios relative to wild-type virus. Further, the DAS181 resistance phenotype was unstable and was substantially reversed over time upon DAS181 withdrawal. In mice, the DAS181-selected viruses exhibited no greater virulence than their wild-type counterparts. Genotypic and phenotypic analysis of DAS181-selected viruses revealed mutations in the haemagglutinin (HA) and neuraminidase (NA) molecules and also changes in HA and NA function.
Conclusions: results indicate that resistance to DAS181 is minimal and unstable. The DAS181-selected IFV isolates exhibit reduced fitness in vitro, likely due to altered HA and NA functions.
Figures
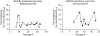
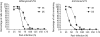


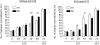
References
-
- WHO. Seasonal Influenza Factsheet. http://www.who.int/mediacentre/factsheets/fs211/en/index.html , 12 July 2010, date last accessed.
-
- Dharan NJ, Gubareva LV, Meyer JJ, et al. Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States. JAMA. 2009;301:1034–41. doi:10.1001/jama.2009.294. - DOI - PubMed
-
- Hatakeyama S, Sugaya N, Ito M, et al. Emergence of influenza B viruses with reduced sensitivity to neuraminidase inhibitors. JAMA. 2007;297:1435–42. doi:10.1001/jama.297.13.1435. - DOI - PubMed
-
- Hayden FG, Hay AJ. Emergence and transmission of influenza A viruses resistant to amantadine and rimantadine. Curr Top Microbiol Immunol. 1992;176:119–30. - PubMed
-
- Moscona A. Oseltamivir resistance—disabling our influenza defenses. N Engl J Med. 2005;353:2633–6. doi:10.1056/NEJMp058291. - DOI - PubMed

