Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist
- PMID: 21097933
- PMCID: PMC3058422
- DOI: 10.1126/science.1197410
Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist
Abstract
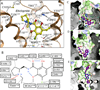
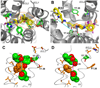
Dopamine modulates movement, cognition, and emotion through activation of dopamine G protein-coupled receptors in the brain. The crystal structure of the human dopamine D3 receptor (D3R) in complex with the small molecule D2R/D3R-specific antagonist eticlopride reveals important features of the ligand binding pocket and extracellular loops. On the intracellular side of the receptor, a locked conformation of the ionic lock and two distinctly different conformations of intracellular loop 2 are observed. Docking of R-22, a D3R-selective antagonist, reveals an extracellular extension of the eticlopride binding site that comprises a second binding pocket for the aryl amide of R-22, which differs between the highly homologous D2R and D3R. This difference provides direction to the design of D3R-selective agents for treating drug abuse and other neuropsychiatric indications.
Figures


References
-
- Civelli O. In: Psychopharmacology : the fourth generation of progress. Bloom F, Kufper D, editors. NY: Raven press; 1995. pp. 155–161.
-
- Levant B. Pharmacol. Rev. 1997;49:231. - PubMed
-
- Sokoloff P, Giros B, Martres M, Bouthenet M, Schwartz J. Nature. 1990;347:146. - PubMed
-
- Shi L, Javitch JA. Annu. Rev. Pharmacol. Toxicol. 2002;42:437. - PubMed
-
- Sokoloff P, et al. Eur. J. Pharmacol. 1992;225:331. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- Y1-GM-1104/GM/NIGMS NIH HHS/United States
- R01 MH054137/MH/NIMH NIH HHS/United States
- DA023694/DA/NIDA NIH HHS/United States
- DA022413/DA/NIDA NIH HHS/United States
- R21 RR025336/RR/NCRR NIH HHS/United States
- R00 DA023694/DA/NIDA NIH HHS/United States
- GM075915/GM/NIGMS NIH HHS/United States
- U54 GM074961/GM/NIGMS NIH HHS/United States
- P50 GM073197/GM/NIGMS NIH HHS/United States
- U54 GM094618/GM/NIGMS NIH HHS/United States
- MH54137/MH/NIMH NIH HHS/United States
- K05 DA022413/DA/NIDA NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- R01 GM089857/GM/NIGMS NIH HHS/United States
- Y1-CO-1020/CO/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

