Modeling the self-assembly of the cellulosome enzyme complex
- PMID: 21098021
- PMCID: PMC3037675
- DOI: 10.1074/jbc.M110.186031
Modeling the self-assembly of the cellulosome enzyme complex
Abstract
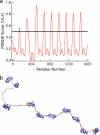
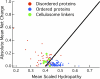
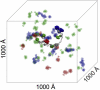
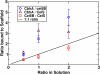
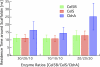
Most bacteria use free enzymes to degrade plant cell walls in nature. However, some bacteria have adopted a different strategy wherein enzymes can either be free or tethered on a protein scaffold forming a complex called a cellulosome. The study of the structure and mechanism of these large macromolecular complexes is an active and ongoing research topic, with the goal of finding ways to improve biomass conversion using cellulosomes. Several mechanisms involved in cellulosome formation remain unknown, including how cellulosomal enzymes assemble on the scaffoldin and what governs the population of cellulosomes created during self-assembly. Here, we present a coarse-grained model to study the self-assembly of cellulosomes. The model captures most of the physical characteristics of three cellulosomal enzymes (Cel5B, CelS, and CbhA) and the scaffoldin (CipA) from Clostridium thermocellum. The protein structures are represented by beads connected by restraints to mimic the flexibility and shapes of these proteins. From a large simulation set, the assembly of cellulosomal enzyme complexes is shown to be dominated by their shape and modularity. The multimodular enzyme, CbhA, binds statistically more frequently to the scaffoldin than CelS or Cel5B. The enhanced binding is attributed to the flexible nature and multimodularity of this enzyme, providing a longer residence time around the scaffoldin. The characterization of the factors influencing the cellulosome assembly process may enable new strategies to create designers cellulosomes.
Figures

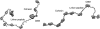



Similar articles
-
Insights into higher-order organization of the cellulosome revealed by a dissect-and-build approach: crystal structure of interacting Clostridium thermocellum multimodular components.J Mol Biol. 2010 Mar 5;396(4):833-9. doi: 10.1016/j.jmb.2010.01.015. Epub 2010 Jan 11. J Mol Biol. 2010. PMID: 20070943
-
Dramatic performance of Clostridium thermocellum explained by its wide range of cellulase modalities.Sci Adv. 2016 Feb 5;2(2):e1501254. doi: 10.1126/sciadv.1501254. eCollection 2016 Feb. Sci Adv. 2016. PMID: 26989779 Free PMC article.
-
In vitro reconstitution of the complete Clostridium thermocellum cellulosome and synergistic activity on crystalline cellulose.Appl Environ Microbiol. 2012 Jun;78(12):4301-7. doi: 10.1128/AEM.07959-11. Epub 2012 Apr 20. Appl Environ Microbiol. 2012. PMID: 22522677 Free PMC article.
-
The Clostridium cellulovorans cellulosome: an enzyme complex with plant cell wall degrading activity.Chem Rec. 2001;1(1):24-32. doi: 10.1002/1528-0691(2001)1:1<24::AID-TCR5>3.0.CO;2-W. Chem Rec. 2001. PMID: 11893054 Review.
-
Cellulosomes-structure and ultrastructure.J Struct Biol. 1998 Dec 15;124(2-3):221-34. doi: 10.1006/jsbi.1998.4065. J Struct Biol. 1998. PMID: 10049808 Review.
Cited by
-
Draft Genomes of Six Philippine Erwinia mallotivora Isolates: Comparative Genomics and Genome-Wide Analysis of Candidate Secreted Proteins.Curr Microbiol. 2022 Apr 18;79(6):164. doi: 10.1007/s00284-022-02857-x. Curr Microbiol. 2022. PMID: 35435500
-
Mapping the deformability of natural and designed cellulosomes in solution.Biotechnol Biofuels Bioprod. 2022 Jun 20;15(1):68. doi: 10.1186/s13068-022-02165-3. Biotechnol Biofuels Bioprod. 2022. PMID: 35725490 Free PMC article.
-
Characterisation of the Effect of the Spatial Organisation of Hemicellulases on the Hydrolysis of Plant Biomass Polymer.Int J Mol Sci. 2020 Jun 19;21(12):4360. doi: 10.3390/ijms21124360. Int J Mol Sci. 2020. PMID: 32575393 Free PMC article.
-
Substrate-Related Factors Affecting Cellulosome-Induced Hydrolysis for Lignocellulose Valorization.Int J Mol Sci. 2019 Jul 8;20(13):3354. doi: 10.3390/ijms20133354. Int J Mol Sci. 2019. PMID: 31288425 Free PMC article. Review.
-
Microbiology: Break down the walls.Nature. 2013 Jan 3;493(7430):36-7. doi: 10.1038/493036a. Nature. 2013. PMID: 23282361 No abstract available.
References
-
- Bayer E. A., Belaich J. P., Shoham Y., Lamed R. (2004) Annu. Rev. Microbiol. 58, 521–554 - PubMed
-
- Doi R. H., Kosugi A. (2004) Nat. Rev. Microbiol. 2, 541–551 - PubMed
-
- Fierobe H. P., Mingardon F., Mechaly A., Bélaïch A., Rincon M. T., Pagès S., Lamed R., Tardif C., Bélaïch J. P., Bayer E. A. (2005) J. Biol. Chem. 280, 16325–16334 - PubMed
-
- Lamed R., Bayer E. A. (1988) Adv. Appl. Microbiol. 33, 1–46
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

