Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes
- PMID: 21098664
- PMCID: PMC3003019
- DOI: 10.1073/pnas.1016147107
Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes
Abstract
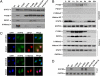
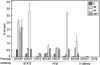
Following its tyrosine phosphorylation, STAT3 is methylated on K140 by the histone methyl transferase SET9 and demethylated by LSD1 when it is bound to a subset of the promoters that it activates. Methylation of K140 is a negative regulatory event, because its blockade greatly increases the steady-state amount of activated STAT3 and the expression of many (i.e., SOCS3) but not all (i.e., CD14) STAT3 target genes. Biological relevance is shown by the observation that overexpression of SOCS3 when K140 cannot be methylated blocks the ability of cells to activate STAT3 in response to IL-6. K140 methylation does not occur with mutants of STAT3 that do not enter nuclei or bind to DNA. Following treatment with IL-6, events at the SOCS3 promoter occur in an ordered sequence, as shown by chromatin immunoprecipitations. Y705-phosphoryl-STAT3 binds first and S727 is then phosphorylated, followed by the coincident binding of SET9 and dimethylation of K140, and lastly by the binding of LSD1. We conclude that the lysine methylation of promoter-bound STAT3 leads to biologically important down-regulation of the dependent responses and that SET9, which is known to help provide an activating methylation mark to H3K4, is recruited to the newly activated SOCS3 promoter by STAT3.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

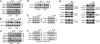
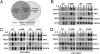

References
-
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. - PubMed
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
-
- Huang J, Berger SL. The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev. 2008;18:152–158. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

