Transspecies dimorphic allelic lineages of the proteasome subunit beta-type 8 gene (PSMB8) in the teleost genus Oryzias
- PMID: 21098669
- PMCID: PMC3003058
- DOI: 10.1073/pnas.1012881107
Transspecies dimorphic allelic lineages of the proteasome subunit beta-type 8 gene (PSMB8) in the teleost genus Oryzias
Abstract
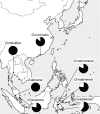
The proteasome subunit β-type 8 (PSMB8) gene in the jawed vertebrate MHC genomic region encodes a catalytic subunit of the immunoproteasome involved in the generation of peptides to be presented by the MHC class I molecules. A teleost, the medaka (Oryzias latipes), has highly diverged dimorphic allelic lineages of the PSMB8 gene with only about 80% amino acid identity, termed "PSMB8d" and "PSMB8N," which have been retained by most wild populations analyzed. To elucidate the evolutionary origin of these two allelic lineages, seven species of the genus Oryzias were analyzed for their PSMB8 allelic sequences using a large number of individuals from wild populations. All the PSMB8 alleles of these species were classified into one of these two allelic lineages based on their nucleotide sequences of exons and introns, indicating that the Oryzias PSMB8 gene has a truly dichotomous allelic lineage. Retention of both allelic lineages was confirmed except for one species. The PSMB8d lineage showed a higher frequency than the PSMB8N lineage in all seven species. The two allelic lineages showed curious substitutions at the 31st and 53rd residues of the mature peptide, probably involved in formation of the S1 pocket, suggesting that these allelic lineages show a functional difference in cleavage specificity. These results indicate that the PSMB8 dimorphism was established before speciation within the genus Oryzias and has been maintained for more than 30-60 million years under a strict and asymmetric balancing selection through several speciation events.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Klein J, Sato A, Nikolaidis N. MHC, TSP, and the origin of species: From immunogenetics to evolutionary genetics. Annu Rev Genet. 2007;41:281–304. - PubMed
-
- Hedrick PW. Genetic polymorphism in heterogeneous environments: The age of genomics. Annu Rev Ecol Evol Syst. 2006;37:67–93.
-
- Figueroa F, Günther E, Klein J. MHC polymorphism pre-dating speciation. Nature. 1988;335:265–267. - PubMed
-
- Lawlor DA, Ward FE, Ennis PD, Jackson AP, Parham P. HLA-A and B polymorphisms predate the divergence of humans and chimpanzees. Nature. 1988;335:268–271. - PubMed
-
- Hughes AL, Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988;335:167–170. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

