Callose synthase GSL7 is necessary for normal phloem transport and inflorescence growth in Arabidopsis
- PMID: 21098675
- PMCID: PMC3075753
- DOI: 10.1104/pp.110.166330
Callose synthase GSL7 is necessary for normal phloem transport and inflorescence growth in Arabidopsis
Abstract
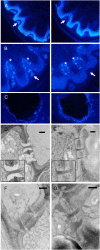
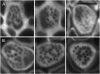
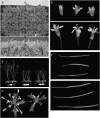
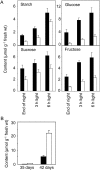
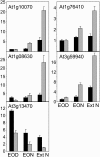
One isoform of callose synthase, Glucan Synthase-Like7 (GSL7), is tightly coexpressed with two isoforms of sucrose synthase (SUS5 and SUS6) known to be confined to phloem sieve elements in Arabidopsis (Arabidopsis thaliana). Investigation of the phenotype of gsl7 mutants of Arabidopsis revealed that the sieve plate pores of stems and roots lack the callose lining seen in wild-type plants. Callose synthesis in other tissues of the plant appears to be unaffected. Although gsl7 plants show only minor phenotypic alterations during vegetative growth, flowering stems are reduced in height and all floral parts are smaller than those of wild-type plants. Several lines of evidence suggest that the reduced growth of the inflorescence is a result of carbohydrate starvation. Levels of sucrose, hexoses, and starch are lower in the terminal bud clusters of gsl7 than in those of wild-type plants. Transcript levels of "starvation" genes expressed in response to low sugars are elevated in the terminal bud clusters of gsl7 plants, at the end of the night, and during an extended night. Pulse-chase experiments with (14)CO(2) show that transport of assimilate in the flowering stem is much slower in gsl7 mutants than in wild-type plants. We suggest that the callose lining of sieve plate pores is essential for normal phloem transport because it confers favorable flow characteristics on the pores.
Figures





References
-
- Arteca RN, Arteca JM. (2000) A novel method for growing Arabidopsis thaliana plants hydroponically. Physiol Plant 108: 188–193
-
- Baud S, Vaultier MN, Rochat C. (2004) Structure and expression profile of the sucrose synthase multigene family in Arabidopsis. J Exp Bot 55: 397–409 - PubMed
-
- Bieniawska Z, Barratt DHP, Garlick AP, Thole V, Kruger NJ, Martin C, Zrenner R, Smith AM. (2007) Analysis of the sucrose synthase gene family in Arabidopsis. Plant J 49: 810–828 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

