Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3
- PMID: 21098734
- PMCID: PMC3015117
- DOI: 10.1105/tpc.110.078964
Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3
Abstract
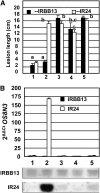
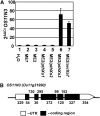
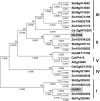
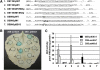
The rice (Oryza sativa) gene xa13 is a recessive resistance allele of Os-8N3, a member of the NODULIN3 (N3) gene family, located on rice chromosome 8. Os-8N3 is a susceptibility (S) gene for Xanthomonas oryzae pv oryzae, the causal agent of bacterial blight, and the recessive allele is defeated by strains of the pathogen producing any one of the type III effectors AvrXa7, PthXo2, or PthXo3, which are all members of the transcription activator-like (TAL) effector family. Both AvrXa7 and PthXo3 induce the expression of a second member of the N3 gene family, here named Os-11N3. Insertional mutagenesis or RNA-mediated silencing of Os-11N3 resulted in plants with loss of susceptibility specifically to strains of X. oryzae pv oryzae dependent on AvrXa7 or PthXo3 for virulence. We further show that AvrXa7 drives expression of Os-11N3 and that AvrXa7 interacts and binds specifically to an effector binding element within the Os-11N3 promoter, lending support to the predictive models for TAL effector binding specificity. The result indicates that variations in the TAL effector repetitive domains are driven by selection to overcome both dominant and recessive forms of resistance to bacterial blight in rice. The finding that Os-8N3 and Os-11N3 encode closely related proteins also provides evidence that N3 proteins have a specific function in facilitating bacterial blight disease.
Figures





References
-
- Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., Lahaye T., Nickstadt A., Bonas U. (2009). Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326: 1509–1512 - PubMed
-
- Chisholm S.T., Coaker G., Day B., Staskawicz B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124: 803–814 - PubMed
-
- Chu C.G., Faris J.D., Xu S.S., Friesen T.L. (2010). Genetic analysis of disease susceptibility contributed by the compatible Tsn1-SnToxA and Snn1-SnTox1 interactions in the wheat-Stagonospora nodorum pathosystem. Theor. Appl. Genet. 120: 1451–1459 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

