The poly A polymerase Star-PAP controls 3'-end cleavage by promoting CPSF interaction and specificity toward the pre-mRNA
- PMID: 21102410
- PMCID: PMC3018792
- DOI: 10.1038/emboj.2010.287
The poly A polymerase Star-PAP controls 3'-end cleavage by promoting CPSF interaction and specificity toward the pre-mRNA
Abstract
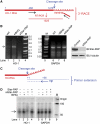
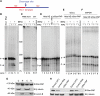
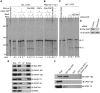
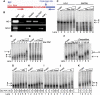
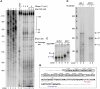
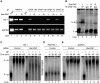
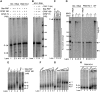
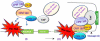
Star-PAP is a poly (A) polymerase (PAP) that is putatively required for 3'-end cleavage and polyadenylation of a select set of pre-messenger RNAs (mRNAs), including heme oxygenase (HO-1) mRNA. To investigate the underlying mechanism, the cleavage and polyadenylation of pre-mRNA was reconstituted with nuclear lysates. siRNA knockdown of Star-PAP abolished cleavage of HO-1, and this phenotype could be rescued by recombinant Star-PAP but not PAPα. Star-PAP directly associated with cleavage and polyadenylation specificity factor (CPSF) 160 and 73 subunits and also the targeted pre-mRNA. In vitro and in vivo Star-PAP was required for the stable association of CPSF complex to pre-mRNA and then CPSF 73 specifically cleaved the mRNA at the 3'-cleavage site. This mechanism is distinct from canonical PAPα, which is recruited to the cleavage complex by interacting with CPSF 160. The data support a model where Star-PAP binds to the RNA, recruits the CPSF complex to the 3'-end of pre-mRNA and then defines cleavage by CPSF 73 and subsequent polyadenylation of its target mRNAs.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Comment in
-
More than one way to make a tail.EMBO J. 2010 Dec 15;29(24):4066-7. doi: 10.1038/emboj.2010.306. EMBO J. 2010. PMID: 21157480 Free PMC article.
References
-
- Barabino SM, Hubner W, Jenny A, Minvielle-Sebastia L, Keller W (1997) The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev 11: 1703–1716 - PubMed
-
- Bardwell VJ, Wickens M, Bienroth S, Keller W, Sproat BS, Lamond AI (1991) Site-directed ribose methylation identifies 2′-OH groups in polyadenylation substrates critical for AAUAAA recognition and poly(A) addition. Cell 65: 125–133 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

