Transforming growth factor-β regulates the sphere-initiating stem cell-like feature in breast cancer through miRNA-181 and ATM
- PMID: 21102523
- PMCID: PMC3063856
- DOI: 10.1038/onc.2010.531
Transforming growth factor-β regulates the sphere-initiating stem cell-like feature in breast cancer through miRNA-181 and ATM
Abstract
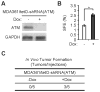
Recent studies indicate that a subset of cancer cells possessing stem cell properties, referred to as cancer-initiating or cancer stem cells (CSCs), have crucial roles in tumor initiation, metastasis and resistance to anticancer therapies. Transforming growth factor (TGF)-β and their family members have been implicated in both normal (embryonic and somatic) stem cells and CSCs. In this study, we observed that exposure to TGF-β increased the population of breast cancer (BC) cells that can form mammospheres in suspension, a feature endowed by stem cells. This was mediated by the micro (mi)RNA family miR-181, which was upregulated by TGF-β at the post-transcriptional level. Levels of the miR-181 family members were elevated in mammospheres grown in undifferentiating conditions, compared with cells grown in two-dimensional conditions. Ataxia telangiectasia mutated (ATM), a target gene of miR-181, exhibited reduced expression in mammospheres and upon TGF-β treatment. Overexpression of miR-181a/b, or depletion of ATM or its substrate CHK2, was sufficient to induce sphere formation in BC cells. Finally, knockdown of ATM enhanced in vivo tumorigenesis of the MDA361 BC cells. Our results elucidate a novel mechanism through which the TGF-β pathway regulates the CSC property by interfering with the tumor suppressor ATM, providing insights into the cellular and environmental factors regulating CSCs, which may guide future studies on therapeutic strategies targeting these cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Ataxia telangiectasia mutated (ATM) inhibition transforms human mammary gland epithelial cells.J Biol Chem. 2010 Apr 23;285(17):13092-106. doi: 10.1074/jbc.M109.078360. Epub 2010 Feb 22. J Biol Chem. 2010. PMID: 20177072 Free PMC article.
-
Breast cancer stem cell-like cells are more sensitive to ionizing radiation than non-stem cells: role of ATM.PLoS One. 2012;7(11):e50423. doi: 10.1371/journal.pone.0050423. Epub 2012 Nov 21. PLoS One. 2012. PMID: 23185620 Free PMC article.
-
miR-18a impairs DNA damage response through downregulation of ataxia telangiectasia mutated (ATM) kinase.PLoS One. 2011;6(9):e25454. doi: 10.1371/journal.pone.0025454. Epub 2011 Sep 27. PLoS One. 2011. PMID: 21980462 Free PMC article.
-
Resistance to DNA-damaging treatment in non-small cell lung cancer tumor-initiating cells involves reduced DNA-PK/ATM activation and diminished cell cycle arrest.Cell Death Dis. 2013 Jan 31;4(1):e478. doi: 10.1038/cddis.2012.211. Cell Death Dis. 2013. PMID: 23370278 Free PMC article.
-
Activation and regulation of ATM kinase activity in response to DNA double-strand breaks.Oncogene. 2007 Dec 10;26(56):7741-8. doi: 10.1038/sj.onc.1210872. Oncogene. 2007. PMID: 18066086 Review.
Cited by
-
MALAT1 promotes gastric adenocarcinoma through the MALAT1/miR-181a-5p/AKT3 axis.Open Biol. 2019 Sep 27;9(9):190095. doi: 10.1098/rsob.190095. Epub 2019 Sep 4. Open Biol. 2019. Retraction in: Open Biol. 2023 Nov;13(11):230404. doi: 10.1098/rsob.230404. PMID: 31480991 Free PMC article. Retracted.
-
TGF-β Induction of miR-143/145 Is Associated to Exercise Response by Influencing Differentiation and Insulin Signaling Molecules in Human Skeletal Muscle.Cells. 2021 Dec 7;10(12):3443. doi: 10.3390/cells10123443. Cells. 2021. PMID: 34943951 Free PMC article.
-
Cancer stem cells and oral cancer: insights into molecular mechanisms and therapeutic approaches.Cancer Cell Int. 2020 Apr 7;20:113. doi: 10.1186/s12935-020-01192-0. eCollection 2020. Cancer Cell Int. 2020. PMID: 32280305 Free PMC article. Review.
-
MicroRNAs: novel players in cancer diagnosis and therapies.Biomed Res Int. 2014;2014:959461. doi: 10.1155/2014/959461. Epub 2014 Jul 2. Biomed Res Int. 2014. PMID: 25101302 Free PMC article. Review.
-
Low dose IR-induced IGF-1-sCLU expression: a p53-repressed expression cascade that interferes with TGFβ1 signaling to confer a pro-survival bystander effect.Oncogene. 2013 Jan 24;32(4):479-90. doi: 10.1038/onc.2012.64. Epub 2012 Mar 5. Oncogene. 2013. PMID: 22391565 Free PMC article.
References
-
- Ailles LE, Weissman IL. Cancer stem cells in solid tumors. Curr Opin Biotechnol. 2007;18:460–6. - PubMed
-
- Austen B, Skowronska A, Baker C, Powell JE, Gardiner A, Oscier D, et al. Mutation status of the residual ATM allele is an important determinant of the cellular response to chemotherapy and survival in patients with chronic lymphocytic leukemia containing an 11q deletion. J Clin Oncol. 2007;25:5448–57. - PubMed
-
- Cicalese A, Bonizzi G, Pasi CE, Faretta M, Ronzoni S, Giulini B, et al. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 2009;138:1083–95. - PubMed
-
- Cordenonsi M, Dupont S, Maretto S, Insinga A, Imbriano C, Piccolo S. Links between tumor suppressors: p53 is required for TGF-beta gene responses by cooperating with Smads. Cell. 2003;113:301–14. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

