Synonymous but not the same: the causes and consequences of codon bias
- PMID: 21102527
- PMCID: PMC3074964
- DOI: 10.1038/nrg2899
Synonymous but not the same: the causes and consequences of codon bias
Abstract
Despite their name, synonymous mutations have significant consequences for cellular processes in all taxa. As a result, an understanding of codon bias is central to fields as diverse as molecular evolution and biotechnology. Although recent advances in sequencing and synthetic biology have helped to resolve longstanding questions about codon bias, they have also uncovered striking patterns that suggest new hypotheses about protein synthesis. Ongoing work to quantify the dynamics of initiation and elongation is as important for understanding natural synonymous variation as it is for designing transgenes in applied contexts.
Figures
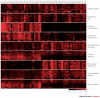
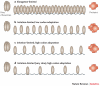
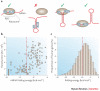

References
-
- Zuckerkandl E, Pauling L. Molecules as documents of evolutionary history. J Theor Biol. 1965;8:357–66. - PubMed
-
- Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346–53. - PubMed
-
- Hershberg R, Petrov DA. Selection on codon bias. Annu Rev Genet. 2008;42:287–99. - PubMed
-
- Chamary JV, Parmley JL, Hurst LD. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat Rev Genet. 2006;7:98–108. - PubMed
-
- Duret L. Evolution of synonymous codon usage in metazoans. Curr Opin Genet Dev. 2002;12:640–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

