Proline residues link the active site to transmembrane domain movements in human nucleoside triphosphate diphosphohydrolase 3 (NTPDase3)
- PMID: 21103216
- PMCID: PMC2947659
- DOI: 10.1007/s11302-010-9180-0
Proline residues link the active site to transmembrane domain movements in human nucleoside triphosphate diphosphohydrolase 3 (NTPDase3)
Abstract
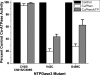
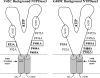
The active sites of the membrane-bound nucleoside triphosphate diphosphohydrolases (NTPDases) regulate and are regulated by coordinated and spatially distant movements of their transmembrane helices, modulating enzyme activity, and substrate specificity. Using site-directed mutagenesis, the roles of the conserved proline residues (N-terminal: P52 and P53; C-terminal: P472, P476, P481, P484, and P485) of human NTPDase3, located in the "linker regions" that connect the N- and C-terminal transmembrane helices with the extracellular active site, were examined. Single cysteine substitutions were strategically placed in the transmembrane domain (N-terminal helix: V42C; C-terminal helix: G489C) to serve as cross-linking "sensors" of helical interactions. These "sensor" background mutant proteins (V42C and G489C NTPDase3) are enzymatically active and are cross-linked by copper phenanthroline less efficiently in the presence of adenosine triphosphate (ATP). Proline to alanine substitutions at P53, P481, P484, and P485 in the V42C background, as well as P53, P481, and P484 in the G489C background, exhibited decreased nucleotidase activities. More importantly, alanine substitutions at P53 and P481 in the V42C background and P481 in the G489C background no longer exhibited the ATP-induced decrease in transmembrane cross-linking efficiency. Interestingly, the P485A mutation abolished oxidative cross-linking at G489C both in the presence and absence of ATP. Taken together, these results suggest a role for proline residues 53 and 481 in the linker regions of human NTPDase3 for coupling nucleotide binding at the enzyme active site to movements and/or rearrangements of the transmembrane helices necessary for optimal nucleotide hydrolysis.
Keywords: Conserved proline residues; Ecto-nucleotidase; Linker region; NTPDase3; Site-directed mutagenesis; Transmembrane cross-linking.
Figures

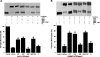

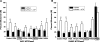
Similar articles
-
Conserved polar residues stabilize transmembrane domains and promote oligomerization in human nucleoside triphosphate diphosphohydrolase 3.Biochemistry. 2009 Oct 13;48(40):9437-47. doi: 10.1021/bi900909g. Biochemistry. 2009. PMID: 19743837 Free PMC article.
-
Conserved lysine 79 is important for activity of ecto-nucleoside triphosphate diphosphohydrolase 3 (NTPDase3).Purinergic Signal. 2004 Dec;1(1):51-8. doi: 10.1007/s11302-004-4741-8. Purinergic Signal. 2004. PMID: 18404400 Free PMC article.
-
Asparagine 81, an invariant glycosylation site near apyrase conserved region 1, is essential for full enzymatic activity of ecto-nucleoside triphosphate diphosphohydrolase 3.Arch Biochem Biophys. 2003 May 1;413(1):107-15. doi: 10.1016/s0003-9861(03)00084-5. Arch Biochem Biophys. 2003. PMID: 12706347
-
Emerging issues of connexin channels: biophysics fills the gap.Q Rev Biophys. 2001 Aug;34(3):325-472. doi: 10.1017/s0033583501003705. Q Rev Biophys. 2001. PMID: 11838236 Review.
-
Therapeutic potentials of ecto-nucleoside triphosphate diphosphohydrolase, ecto-nucleotide pyrophosphatase/phosphodiesterase, ecto-5'-nucleotidase, and alkaline phosphatase inhibitors.Med Res Rev. 2014 Jul;34(4):703-43. doi: 10.1002/med.21302. Epub 2013 Sep 23. Med Res Rev. 2014. PMID: 24115166 Review.
Cited by
-
Cellular function and molecular structure of ecto-nucleotidases.Purinergic Signal. 2012 Sep;8(3):437-502. doi: 10.1007/s11302-012-9309-4. Epub 2012 May 4. Purinergic Signal. 2012. PMID: 22555564 Free PMC article. Review.
-
Fusion of a proline-rich oligopeptide to the C-terminus of a ruminal xylanase improves catalytic efficiency.Bioengineered. 2022 Apr;13(4):10482-10492. doi: 10.1080/21655979.2022.2061290. Bioengineered. 2022. PMID: 35441569 Free PMC article.
-
The GDA1_CD39 superfamily: NTPDases with diverse functions.Purinergic Signal. 2011 Mar;7(1):21-45. doi: 10.1007/s11302-010-9214-7. Epub 2011 Jan 21. Purinergic Signal. 2011. PMID: 21484095 Free PMC article.
-
Pathologic complete response to neoadjuvant imatinib of a gastric stromal tumor with concomitant mutations in KIT: A case report and literature review.Clin Case Rep. 2023 Jun 7;11(6):e7463. doi: 10.1002/ccr3.7463. eCollection 2023 Jun. Clin Case Rep. 2023. PMID: 37305871 Free PMC article.
-
Current status and proposed roles for nitric oxide as a key mediator of the effects of extracellular nucleotides on plant growth.Front Plant Sci. 2013 Oct 25;4:427. doi: 10.3389/fpls.2013.00427. Front Plant Sci. 2013. PMID: 24298275 Free PMC article. Review.
References
-
- Murphy-Piedmonte DM, Crawford PA, Kirley TL. Bacterial expression, folding, purification and characterization of soluble NTPDase5 (CD39L4) ecto-nucleotidase. Biochim Biophys Acta. 2005;1747:251–259. - PubMed
-
- Stout JG, Strobel RS, Kirley TL. Identification and immunolocalization of ecto-ATPDase in chicken stomach. Biochem Mol Biol Int. 1995;36:529–535. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

