Tyr(682) in the intracellular domain of APP regulates amyloidogenic APP processing in vivo
- PMID: 21103325
- PMCID: PMC2982846
- DOI: 10.1371/journal.pone.0015503
Tyr(682) in the intracellular domain of APP regulates amyloidogenic APP processing in vivo
Abstract
Background: The pathogenesis of Alzheimer's disease is attributed to misfolding of Amyloid-β (Aβ) peptides. Aβ is generated during amyloidogenic processing of Aβ-precursor protein (APP). Another characteristic of the AD brain is increased phosphorylation of APP amino acid Tyr(682). Tyr(682) is part of the Y(682)ENPTY(687) motif, a docking site for interaction with cytosolic proteins that regulate APP metabolism and signaling. For example, normal Aβ generation and secretion are dependent upon Tyr(682) in vitro. However, physiological functions of Tyr(682) are unknown.
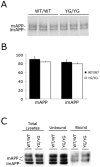
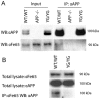
Methodology/principal findings: To this end, we have generated an APP Y682G knock-in (KI) mouse to help dissect the role of APP Tyr(682) in vivo. We have analyzed proteolytic products from both the amyloidogenic and non-amyloidogenic processing of APP and measure a profound shift towards non-amyloidogenic processing in APP KI mice. In addition, we demonstrate the essential nature of amino acid Tyr(682) for the APP/Fe65 interaction in vivo.
Conclusions/significance: Together, these observations point to an essential role of APP intracellular domain for normal APP processing and function in vivo, and provide rationale for further studies into physiological functions associated with this important phosphorylation site.
Conflict of interest statement
Figures
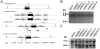
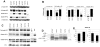
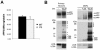


References
-
- Hardy JA, Higgins GA. Alzheimer's disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. - PubMed
-
- Selkoe DJ. The genetics and molecular pathology of Alzheimer's disease: roles of amyloid and the presenilins. Neurol Clin. 2000;18:903–922. - PubMed
-
- Passer B, Pellegrini L, Russo C, Siegel RM, Lenardo MJ, et al. Generation of an apoptotic intracellular peptide by gamma-secretase cleavage of Alzheimer's amyloid beta protein precursor. J Alzheimers Dis. 2000;2:289–301. - PubMed
-
- Cao X, Sudhof TC. A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293:115–120. - PubMed
-
- Stokin GB, Lillo C, Falzone TL, Brusch RG, Rockenstein E, et al. Axonopathy and transport deficits early in the pathogenesis of Alzheimer's disease. Science. 2005;307:1282–1288. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

