Genetic analyses of interactions among gibberellin, abscisic acid, and brassinosteroids in the control of flowering time in Arabidopsis thaliana
- PMID: 21103336
- PMCID: PMC2984439
- DOI: 10.1371/journal.pone.0014012
Genetic analyses of interactions among gibberellin, abscisic acid, and brassinosteroids in the control of flowering time in Arabidopsis thaliana
Abstract
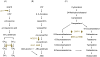
Background: Genetic interactions between phytohormones in the control of flowering time in Arabidopsis thaliana have not been extensively studied. Three phytohormones have been individually connected to the floral-timing program. The inductive function of gibberellins (GAs) is the most documented. Abscisic acid (ABA) has been demonstrated to delay flowering. Finally, the promotive role of brassinosteroids (BRs) has been established. It has been reported that for many physiological processes, hormone pathways interact to ensure an appropriate biological response.
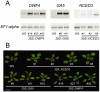
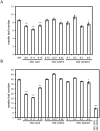
Methodology: We tested possible genetic interactions between GA-, ABA-, and BR-dependent pathways in the control of the transition to flowering. For this, single and double mutants deficient in the biosynthesis of GAs, ABA, and BRs were used to assess the effect of hormone deficiency on the timing of floral transition. Also, plants that over-express genes encoding rate-limiting enzymes in each biosynthetic pathway were generated and the flowering time of these lines was investigated.
Conclusions: Loss-of-function studies revealed a complex relationship between GAs and ABA, and between ABA and BRs, and suggested a cross-regulatory relation between GAs to BRs. Gain-of-function studies revealed that GAs were clearly limiting in their sufficiency of action, whereas increases in BRs and ABA led to a more modest phenotypic effect on floral timing. We conclude from our genetic tests that the effects of GA, ABA, and BR on timing of floral induction are only in partially coordinated action.
Conflict of interest statement
Figures

Similar articles
-
Brassinosteroids Are Master Regulators of Gibberellin Biosynthesis in Arabidopsis.Plant Cell. 2015 Aug;27(8):2261-72. doi: 10.1105/tpc.15.00433. Epub 2015 Aug 4. Plant Cell. 2015. PMID: 26243314 Free PMC article.
-
High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds.Plant Physiol. 2008 Mar;146(3):1368-85. doi: 10.1104/pp.107.113738. Epub 2007 Dec 27. Plant Physiol. 2008. PMID: 18162586 Free PMC article.
-
Pleiotropic effects of the male sterile33 (ms33) mutation in Arabidopsis are associated with modifications in endogenous gibberellins, indole-3-acetic acid and abscisic acid.Planta. 2004 Aug;219(4):649-60. doi: 10.1007/s00425-004-1270-1. Epub 2004 Apr 24. Planta. 2004. PMID: 15107994
-
Gibberellin as a factor in floral regulatory networks.J Exp Bot. 2009;60(7):1979-89. doi: 10.1093/jxb/erp040. Epub 2009 Mar 5. J Exp Bot. 2009. PMID: 19264752 Review.
-
New insights into gibberellin signaling in regulating flowering in Arabidopsis.J Integr Plant Biol. 2020 Jan;62(1):118-131. doi: 10.1111/jipb.12892. Epub 2020 Jan 8. J Integr Plant Biol. 2020. PMID: 31785071 Review.
Cited by
-
Transcription profiles reveal the regulatory mechanisms of spur bud changes and flower induction in response to shoot bending in apple (Malus domestica Borkh.).Plant Mol Biol. 2019 Jan;99(1-2):45-66. doi: 10.1007/s11103-018-0801-2. Epub 2018 Dec 5. Plant Mol Biol. 2019. PMID: 30519825
-
Significant increases in Donghong kiwifruit yield by a novel umbrella-shaped trellis system and identification of associated molecular mechanisms.Front Plant Sci. 2023 Mar 13;14:1143525. doi: 10.3389/fpls.2023.1143525. eCollection 2023. Front Plant Sci. 2023. PMID: 36993843 Free PMC article.
-
Detecting the QTL-allele system conferring flowering date in a nested association mapping population of soybean using a novel procedure.Theor Appl Genet. 2017 Nov;130(11):2297-2314. doi: 10.1007/s00122-017-2960-y. Epub 2017 Aug 10. Theor Appl Genet. 2017. PMID: 28799029
-
Parasitic-Plant Parasite Rewires Flowering Pathways to Induce Stem-Derived Galls.Plant Direct. 2025 Aug 20;9(8):e70099. doi: 10.1002/pld3.70099. eCollection 2025 Aug. Plant Direct. 2025. PMID: 40842965 Free PMC article.
-
Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development.Plant Cell. 2011 Apr;23(4):1219-30. doi: 10.1105/tpc.111.084475. Epub 2011 Apr 19. Plant Cell. 2011. PMID: 21505068 Free PMC article. Review.
References
-
- Bernier G, Perilleux C. A physiological overview of the genetics of flowering time control. Plant Biotechnol J. 2005;3:3–16. - PubMed
-
- Koornneef M, Hanhart CJ, van der Veen JH. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet. 1991;229:57–66. - PubMed
-
- Putterill J, Laurie R, Macknight R. It's time to flower: the genetic control of flowering time. Bioessays. 2004;26:363–373. - PubMed
-
- Davis SJ. Integrating hormones into the floral-transition pathway of Arabidopsis thaliana. Plant Cell Environ. 2009;32:1201–1210. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

