Characterisation of PduS, the pdu metabolosome corrin reductase, and evidence of substructural organisation within the bacterial microcompartment
- PMID: 21103360
- PMCID: PMC2982820
- DOI: 10.1371/journal.pone.0014009
Characterisation of PduS, the pdu metabolosome corrin reductase, and evidence of substructural organisation within the bacterial microcompartment
Abstract
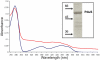
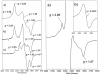
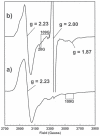
PduS is a corrin reductase and is required for the reactivation of the cobalamin-dependent diol dehydratase. It is one component encoded within the large propanediol utilisation (pdu) operon, which is responsible for the catabolism of 1,2-propanediol within a self-assembled proteinaceous bacterial microcompartment. The enzyme is responsible for the reactivation of the cobalamin coenzyme required by the diol dehydratase. The gene for the cobalamin reductase from Citrobacter freundii (pduS) has been cloned to allow the protein to be overproduced recombinantly in E. coli with an N-terminal His-tag. Purified recombinant PduS is shown to be a flavoprotein with a non-covalently bound FMN that also contains two coupled [4Fe-4S] centres. It is an NADH-dependent flavin reductase that is able to mediate the one-electron reductions of cob(III)alamin to cob(II)alamin and cob(II)alamin to cob(I)alamin. The [4Fe-4S] centres are labile to oxygen and their presence affects the midpoint redox potential of flavin. Evidence is presented that PduS is able to bind cobalamin, which is inconsistent with the view that PduS is merely a flavin reductase. PduS is also shown to interact with one of the shell proteins of the metabolosome, PduT, which is also thought to contain an [Fe-S] cluster. PduS is shown to act as a corrin reductase and its interaction with a shell protein could allow for electron passage out of the bacterial microcompartment.
Conflict of interest statement
Figures


Similar articles
-
Biochemical evidence that the pduS gene encodes a bifunctional cobalamin reductase.Microbiology (Reading). 2005 Apr;151(Pt 4):1169-1177. doi: 10.1099/mic.0.27755-0. Microbiology (Reading). 2005. PMID: 15817784
-
Characterization of the PduS cobalamin reductase of Salmonella enterica and its role in the Pdu microcompartment.J Bacteriol. 2010 Oct;192(19):5071-80. doi: 10.1128/JB.00575-10. Epub 2010 Jul 23. J Bacteriol. 2010. PMID: 20656910 Free PMC article.
-
Structure of PduT, a trimeric bacterial microcompartment protein with a 4Fe-4S cluster-binding site.Acta Crystallogr D Biol Crystallogr. 2011 Feb;67(Pt 2):91-6. doi: 10.1107/S0907444910050201. Epub 2011 Jan 8. Acta Crystallogr D Biol Crystallogr. 2011. PMID: 21245529
-
Reduction of Cob(III)alamin to Cob(II)alamin in Salmonella enterica serovar typhimurium LT2.J Bacteriol. 2000 Aug;182(15):4304-9. doi: 10.1128/JB.182.15.4304-4309.2000. J Bacteriol. 2000. PMID: 10894741 Free PMC article.
-
Lactobacillus reuteri DSM 20016 produces cobalamin-dependent diol dehydratase in metabolosomes and metabolizes 1,2-propanediol by disproportionation.J Bacteriol. 2008 Jul;190(13):4559-67. doi: 10.1128/JB.01535-07. Epub 2008 May 9. J Bacteriol. 2008. PMID: 18469107 Free PMC article.
Cited by
-
Effect of bio-engineering on size, shape, composition and rigidity of bacterial microcompartments.Sci Rep. 2016 Nov 15;6:36899. doi: 10.1038/srep36899. Sci Rep. 2016. PMID: 27845382 Free PMC article.
-
The PduQ enzyme is an alcohol dehydrogenase used to recycle NAD+ internally within the Pdu microcompartment of Salmonella enterica.PLoS One. 2012;7(10):e47144. doi: 10.1371/journal.pone.0047144. Epub 2012 Oct 15. PLoS One. 2012. PMID: 23077559 Free PMC article.
-
Exploring bacterial organelle interactomes: a model of the protein-protein interaction network in the Pdu microcompartment.PLoS Comput Biol. 2015 Feb 3;11(2):e1004067. doi: 10.1371/journal.pcbi.1004067. eCollection 2015 Feb. PLoS Comput Biol. 2015. PMID: 25646976 Free PMC article.
-
Light-Driven Hybrid Nanoreactor Harnessing the Synergy of Carboxysomes and Organic Frameworks for Efficient Hydrogen Production.ACS Catal. 2024 Dec 6;14(24):18603-18614. doi: 10.1021/acscatal.4c03672. eCollection 2024 Dec 20. ACS Catal. 2024. PMID: 39722887 Free PMC article.
-
Electrochemical cofactor recycling of bacterial microcompartments.Proc Natl Acad Sci U S A. 2024 Dec 3;121(49):e2414220121. doi: 10.1073/pnas.2414220121. Epub 2024 Nov 25. Proc Natl Acad Sci U S A. 2024. PMID: 39585991 Free PMC article.
References
-
- Banerjee R, Ragsdale SW. The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes. Annu Rev Biochem. 2003;72:209–247. - PubMed
-
- Roth JR, Lawrence JG, Bobik TA. Cobalamin (coenzyme B12): synthesis and biological significance. Annu Rev Microbiol. 1996;50:137–181. - PubMed
-
- Walker GA, Murphy S, Huennekens FM. Enzymatic conversion of vitamin B 12a to adenosyl-B 12: evidence for the existence of two separate reducing systems. Arch Biochem Biophys. 1969;134:95–102. - PubMed
-
- Warren MJ, Raux E, Schubert HL, Escalante-Semerena JC. The biosynthesis of adenosylcobalamin (vitamin B12). Nat Prod Rep. 2002;19:390–412. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

