SNP selection in genome-wide and candidate gene studies via penalized logistic regression
- PMID: 21104890
- PMCID: PMC3410531
- DOI: 10.1002/gepi.20543
SNP selection in genome-wide and candidate gene studies via penalized logistic regression
Abstract
Penalized regression methods offer an attractive alternative to single marker testing in genetic association analysis. Penalized regression methods shrink down to zero the coefficient of markers that have little apparent effect on the trait of interest, resulting in a parsimonious subset of what we hope are true pertinent predictors. Here we explore the performance of penalization in selecting SNPs as predictors in genetic association studies. The strength of the penalty can be chosen either to select a good predictive model (via methods such as computationally expensive cross validation), through maximum likelihood-based model selection criterion (such as the BIC), or to select a model that controls for type I error, as done here. We have investigated the performance of several penalized logistic regression approaches, simulating data under a variety of disease locus effect size and linkage disequilibrium patterns. We compared several penalties, including the elastic net, ridge, Lasso, MCP and the normal-exponential-γ shrinkage prior implemented in the hyperlasso software, to standard single locus analysis and simple forward stepwise regression. We examined how markers enter the model as penalties and P-value thresholds are varied, and report the sensitivity and specificity of each of the methods. Results show that penalized methods outperform single marker analysis, with the main difference being that penalized methods allow the simultaneous inclusion of a number of markers, and generally do not allow correlated variables to enter the model, producing a sparse model in which most of the identified explanatory markers are accounted for.
© 2010 Wiley-Liss, Inc.
Figures

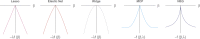
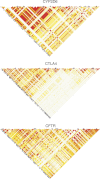


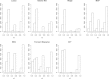
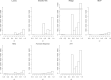
References
-
- Barratt BJ, Payne F, Lowe CE, Hermann R, Healy BC, Harold D, Concannon P, Gharani N, McCarthy MI, Olavensen MG, McCormack R, Guja C, Ionescu-Tirgoviste C, Undlien DE, Ronningen KS, Gillespie KM, Tuomilehto-Wolf E, Tuomilehto J, Benett ST, Clayton DG. Remapping the insulin gene/IDDM2 locus in type 1 diabetes. Diabetes. 2004;53:1884–1889. - PubMed
-
- Breheny P, Huang J. 2008. Penalized methods for bi-level variable selection. Technical Report 393, Department of Statistics and Actuarial Science, University of Iowa.
-
- Breiman L. Better subset regression using the nonnegative garrote. Technometrics. 1995;37:373–384. doi: http://dx.doi.org/10.2307/1269730. - DOI
-
- Breiman L. Bagging predictors. Mach Learn. 1996;24:123–140.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

