Improved Tet-responsive promoters with minimized background expression
- PMID: 21106052
- PMCID: PMC3002914
- DOI: 10.1186/1472-6750-10-81
Improved Tet-responsive promoters with minimized background expression
Abstract
Background: The performance of the tetracycline controlled transcriptional activation system (Tet system) depends critically on the choice of minimal promoters. They are indispensable to warrant low expression levels with the system turned "off". On the other hand, they must support high level of gene expression in the "on"-state.
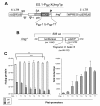
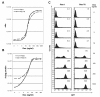
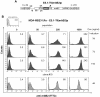
Results: In this study, we systematically modified the widely used Cytomegalovirus (CMV) minimal promoter to further minimize background expression, resulting in an improved dynamic expression range. Using both plasmid-based and retroviral gene delivery, our analysis revealed that especially background expression levels could be significantly reduced when compared to previously established "standard" promoter designs. Our results also demonstrate the possibility to fine-tune expression levels in non-clonal cell populations. They also imply differences regarding the requirements for tight regulation and high level induction between transient and stable gene transfer systems.
Conclusions: Until now, our understanding of mammalian transcriptional regulation including promoter architecture is limited. Nevertheless, the partly empirical modification of cis-elements as shown in this study can lead to the specific improvement of the performance of minimal promoters. The novel composite Ptet promoters introduced here will further expand the utility of the Tet system.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

