The rapid generation of chimerical genes expanding protein diversity in zebrafish
- PMID: 21106061
- PMCID: PMC3091775
- DOI: 10.1186/1471-2164-11-657
The rapid generation of chimerical genes expanding protein diversity in zebrafish
Abstract
Background: Variation of gene number among species indicates that there is a general process of new gene origination. One of the major mechanism providing raw materials for the origin of new genes is gene duplication. Retroposition, as a special type of gene duplication- the RNA-based duplication, has been found to play an important role in new gene evolution in mammals and plants, but little is known about the process in the teleostei genome.
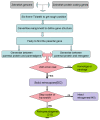
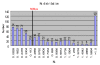
Results: Here we screened the zebrafish genome for identification of retrocopies and new chimerical retrogenes and investigated their origination and evolution. We identified 652 retrocopies, of which 440 are intact retrogenes and 212 are pseudogenes. Retrocopies have long been considered evolutionary dead ends without functional significance due to the presumption that retrocopies lack the regulatory element needed for expression. However, 437 transcribed retrocopies were identified from all of the retrocopies. This discovery combined with the substitution analysis suggested that the majority of all retrocopies are subject to negative selection, indicating that most of the retrocopies may be functional retrogenes. Moreover, we found that 95 chimerical retrogenes had recruited new sequences from neighboring genomic regions that formed de novo splice sites, thus generating new intron-containing chimeric genes. Based on our analysis of 38 pairs of orthologs between Cyprinus carpio and Danio rerio, we found that the synonymous substitution rate of zebrafish genes is 4.13×10⁻⁹ substitution per silent site per year. We also found 10 chimerical retrogenes that were created in the last 10 million years, which is 7.14 times the rate of 0.14 chimerical retrogenes per million years in the primate lineage toward human and 6.25 times the rate of 0.16 chimerical genes per million years in Drosophila. This is among the most rapid rates of generation of chimerical genes, just next to the rice.
Conclusion: There is compelling evidence that much of the extensive transcriptional activity of retrogenes does not represent transcriptional "noise" but indicates the functionality of these retrogenes. Our results indicate that retroposition created a large amount of new genes in the zebrafish genome, which has contributed significantly to the evolution of the fish genome.
Figures

Similar articles
-
Correlated expression of retrocopies and parental genes in zebrafish.Mol Genet Genomics. 2016 Apr;291(2):723-37. doi: 10.1007/s00438-015-1140-5. Epub 2015 Nov 11. Mol Genet Genomics. 2016. PMID: 26561303
-
Genomic exploration of retrocopies in Insect pests of plants and their role in the expansion of heat shock proteins superfamily as evolutionary targets.BMC Genomics. 2024 Nov 20;25(1):1116. doi: 10.1186/s12864-024-11056-w. BMC Genomics. 2024. PMID: 39567882 Free PMC article.
-
LTR-mediated retroposition as a mechanism of RNA-based duplication in metazoans.Genome Res. 2016 Dec;26(12):1663-1675. doi: 10.1101/gr.204925.116. Epub 2016 Oct 20. Genome Res. 2016. PMID: 27934698 Free PMC article.
-
Cancer, Retrogenes, and Evolution.Life (Basel). 2021 Jan 19;11(1):72. doi: 10.3390/life11010072. Life (Basel). 2021. PMID: 33478113 Free PMC article. Review.
-
Not So Dead Genes-Retrocopies as Regulators of Their Disease-Related Progenitors and Hosts.Cells. 2021 Apr 15;10(4):912. doi: 10.3390/cells10040912. Cells. 2021. PMID: 33921034 Free PMC article. Review.
Cited by
-
Foamy-like endogenous retroviruses are extensive and abundant in teleosts.Virus Evol. 2016 Oct 30;2(2):vew032. doi: 10.1093/ve/vew032. eCollection 2016 Jul. Virus Evol. 2016. PMID: 28058112 Free PMC article.
-
Genome-wide identification, characterization, and expression analysis of lineage-specific genes within zebrafish.BMC Genomics. 2013 Jan 31;14:65. doi: 10.1186/1471-2164-14-65. BMC Genomics. 2013. PMID: 23368736 Free PMC article.
-
The Genomic Impact of Gene Retrocopies: What Have We Learned from Comparative Genomics, Population Genomics, and Transcriptomic Analyses?Genome Biol Evol. 2017 Jun 1;9(6):1351-1373. doi: 10.1093/gbe/evx081. Genome Biol Evol. 2017. PMID: 28605529 Free PMC article.
-
AtFusionDB: a database of fusion transcripts in Arabidopsis thaliana.Database (Oxford). 2019 Jan 1;2019:bay135. doi: 10.1093/database/bay135. Database (Oxford). 2019. PMID: 30624648 Free PMC article.
-
Interpopulation differences of retroduplication variations (RDVs) in rice retrogenes and their phenotypic correlations.Comput Struct Biotechnol J. 2021 Jan 5;19:600-611. doi: 10.1016/j.csbj.2020.12.046. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 33510865 Free PMC article.
References
-
- Li W-H. Molecular Evolution. Sunderland, MA.: Sinauer Associates; 1997.
-
- Brosius J. Retroposons--seeds of evolution. Science (New York, NY) 1991;251(4995):753. - PubMed
-
- Long M, Langley CH. Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science (New York, NY) 1993;260(5104):91–95. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

