Evolution of the Kdo2-lipid A biosynthesis in bacteria
- PMID: 21106097
- PMCID: PMC3087551
- DOI: 10.1186/1471-2148-10-362
Evolution of the Kdo2-lipid A biosynthesis in bacteria
Abstract
Background: Lipid A is the highly immunoreactive endotoxic center of lipopolysaccharide (LPS). It anchors the LPS into the outer membrane of most Gram-negative bacteria. Lipid A can be recognized by animal cells, triggers defense-related responses, and causes Gram-negative sepsis. The biosynthesis of Kdo2-lipid A, the LPS substructure, involves with nine enzymatic steps.
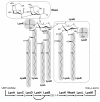
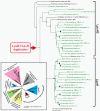
Results: In order to elucidate the evolutionary pathway of Kdo2-lipid A biosynthesis, we examined the distribution of genes encoding the nine enzymes across bacteria. We found that not all Gram-negative bacteria have all nine enzymes. Some Gram-negative bacteria have no genes encoding these enzymes and others have genes only for the first four enzymes (LpxA, LpxC, LpxD, and LpxB). Among the nine enzymes, five appeared to have arisen from three independent gene duplication events. Two of such events happened within the Proteobacteria lineage, followed by functional specialization of the duplicated genes and pathway optimization in these bacteria.
Conclusions: The nine-enzyme pathway, which was established based on the studies mainly in Escherichia coli K12, appears to be the most derived and optimized form. It is found only in E. coli and related Proteobacteria. Simpler and probably less efficient pathways are found in other bacterial groups, with Kdo2-lipid A variants as the likely end products. The Kdo2-lipid A biosynthetic pathway exemplifies extremely plastic evolution of bacterial genomes, especially those of Proteobacteria, and how these mainly pathogenic bacteria have adapted to their environment.
Figures


Similar articles
-
Kdo2 -lipid A: structural diversity and impact on immunopharmacology.Biol Rev Camb Philos Soc. 2015 May;90(2):408-27. doi: 10.1111/brv.12114. Epub 2014 May 16. Biol Rev Camb Philos Soc. 2015. PMID: 24838025 Free PMC article. Review.
-
[Kdo2-lipid A modification in gram-negative bacteria--a review].Wei Sheng Wu Xue Bao. 2013 Feb 4;53(2):111-7. Wei Sheng Wu Xue Bao. 2013. PMID: 23627103 Review. Chinese.
-
Immuno-Stimulatory Activity of Escherichia coli Mutants Producing Kdo2-Monophosphoryl-Lipid A or Kdo2-Pentaacyl-Monophosphoryl-Lipid A.PLoS One. 2015 Dec 28;10(12):e0144714. doi: 10.1371/journal.pone.0144714. eCollection 2015. PLoS One. 2015. PMID: 26710252 Free PMC article.
-
Lipopolysaccharide: Biosynthetic pathway and structure modification.Prog Lipid Res. 2010 Apr;49(2):97-107. doi: 10.1016/j.plipres.2009.06.002. Epub 2009 Oct 6. Prog Lipid Res. 2010. PMID: 19815028 Review.
-
A Complete Pathway Model for Lipid A Biosynthesis in Escherichia coli.PLoS One. 2015 Apr 28;10(4):e0121216. doi: 10.1371/journal.pone.0121216. eCollection 2014. PLoS One. 2015. PMID: 25919634 Free PMC article.
Cited by
-
Manganese Is Essential for PlcP Metallophosphoesterase Activity Involved in Lipid Remodeling in Abundant Marine Heterotrophic Bacteria.Appl Environ Microbiol. 2018 Jul 17;84(15):e01109-18. doi: 10.1128/AEM.01109-18. Print 2018 Aug 1. Appl Environ Microbiol. 2018. PMID: 29802183 Free PMC article.
-
Novel microbiota Mesosutterella faecium sp. nov. has a protective effect against inflammatory bowel disease.Front Microbiol. 2024 Apr 2;15:1342098. doi: 10.3389/fmicb.2024.1342098. eCollection 2024. Front Microbiol. 2024. PMID: 38633706 Free PMC article.
-
Antimicrobial Resistance and in silico Virulence Profiling of Aliarcobacter butzleri Strains From German Water Poultry.Front Microbiol. 2020 Dec 14;11:617685. doi: 10.3389/fmicb.2020.617685. eCollection 2020. Front Microbiol. 2020. PMID: 33381106 Free PMC article.
-
Current Progress in the Structural and Biochemical Characterization of Proteins Involved in the Assembly of Lipopolysaccharide.Int J Microbiol. 2018 Nov 25;2018:5319146. doi: 10.1155/2018/5319146. eCollection 2018. Int J Microbiol. 2018. PMID: 30595696 Free PMC article. Review.
-
Exome and Tissue-Associated Microbiota as Predictive Markers of Response to Neoadjuvant Treatment in Locally Advanced Rectal Cancer.Front Oncol. 2022 Mar 22;12:809441. doi: 10.3389/fonc.2022.809441. eCollection 2022. Front Oncol. 2022. PMID: 35392220 Free PMC article.
References
-
- Ludwig W, Klenk HP. In: Bergey's Manual of Systematic Bacteriology. 2. Boon DR, Castenholz RW, editor. Vol. 1. New York, NY: Springer; 2001. Overview: a phylogenetic backbone and taxonomic framework for prokaryotic systematics; pp. 49–65.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

