Molecular characterization and mRNA expression of two key enzymes of hypoxia-sensing pathways in eastern oysters Crassostrea virginica (Gmelin): hypoxia-inducible factor α (HIF-α) and HIF-prolyl hydroxylase (PHD)
- PMID: 21106446
- PMCID: PMC3102143
- DOI: 10.1016/j.cbd.2010.10.003
Molecular characterization and mRNA expression of two key enzymes of hypoxia-sensing pathways in eastern oysters Crassostrea virginica (Gmelin): hypoxia-inducible factor α (HIF-α) and HIF-prolyl hydroxylase (PHD)
Abstract
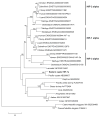
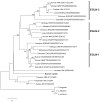
Oxygen homeostasis is crucial for development, survival and normal function of all metazoans. A family of transcription factors called hypoxia-inducible factors (HIF) is critical in mediating the adaptive responses to reduced oxygen availability. The HIF transcription factor consists of a constitutively expressed β subunit and an oxygen-dependent α subunit; the abundance of the latter determines the activity of HIF and is regulated by a family of O(2)- and Fe(2+)-dependent enzymes prolyl hydroxylases (PHDs). Currently very little is known about the function of this important pathway and the molecular structure of its key players in hypoxia-tolerant intertidal mollusks including oysters, which are among the animal champions of anoxic and hypoxic tolerance and thus can serve as excellent models to study the role of HIF cascade in adaptations to oxygen deficiency. We have isolated transcripts of two key components of the oxygen sensing pathway - the oxygen-regulated HIF-α subunit and PHD - from an intertidal mollusk, the eastern oyster Crassostrea virginica, and determined the transcriptional responses of these two genes to anoxia, hypoxia and cadmium (Cd) stress. HIF-α and PHD homologs from eastern oysters C. virginica show significant sequence similarity and share key functional domains with the earlier described isoforms from vertebrates and invertebrates. Phylogenetic analysis shows that genetic diversification of HIF and PHD isoforms occurred within the vertebrate lineage indicating functional diversification and specialization of the oxygen-sensing pathways in this group, which parallels situation observed for many other important genes. HIF-α and PHD homologs are broadly expressed at the mRNA level in different oyster tissues and show transcriptional responses to prolonged hypoxia in the gills consistent with their putative role in oxygen sensing and the adaptive response to hypoxia. Similarity in amino acid sequence, domain structure and transcriptional responses between HIF-α and PHD homologs from oysters and other invertebrate and vertebrate species implies the highly conserved functions of these genes throughout the evolutionary history of animals, in accordance with their critical role in oxygen sensing and homeostasis.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures



References
-
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–52. - PubMed
-
- Appelhoff RJ, Tian Y-M, Raval RR, Turley H, Harris AL, Pugh CW, Ratcliffe PJ, Gleadle JM. Differential Function of the Prolyl Hydroxylases PHD1, PHD2, and PHD3 in the Regulation of Hypoxia-inducible Factor. J Biol Chem. 2004;279:38458–38465. - PubMed
-
- Aprelikova O, Chandramouli GVR, Wood M, Vasselli JR, Riss J, Maranchie JK, Linehan WM, Barrett JC. Regulation of HIF prolyl hydroxylases by hypoxia-inducible factors. J Cell Biochem. 2004;92:491–501. - PubMed
-
- Aragones J, Fraisl P, Baes M, Carmeliet P. Oxygen sensors at the crossroad of metabolism. Cell Metabolism. 2009;9:11–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

