Structures of carbon catabolite protein A-(HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators
- PMID: 21106498
- PMCID: PMC3074128
- DOI: 10.1093/nar/gkq1177
Structures of carbon catabolite protein A-(HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators
Abstract
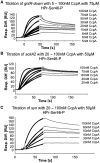
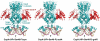
In Gram-positive bacteria, carbon catabolite protein A (CcpA) is the master regulator of carbon catabolite control, which ensures optimal energy usage under diverse conditions. Unlike other LacI-GalR proteins, CcpA is activated for DNA binding by first forming a complex with the phosphoprotein HPr-Ser46-P. Bacillus subtilis CcpA functions as both a transcription repressor and activator and binds to more than 50 operators called catabolite response elements (cres). These sites are highly degenerate with the consensus, WTGNNARCGNWWWCAW. How CcpA-(HPr-Ser46-P) binds such diverse sequences is unclear. To gain insight into this question, we solved the structures of the CcpA-(HPr-Ser46-P) complex bound to three different operators, the synthetic (syn) cre, ackA2 cre and gntR-down cre. Strikingly, the structures show that the CcpA-bound operators display different bend angles, ranging from 31° to 56°. These differences are accommodated by a flexible linkage between the CcpA helix-turn-helix-loop-helix motif and hinge helices, which allows independent docking of these DNA-binding modules. This flexibility coupled with an abundance of non-polar residues capable of non-specific nucleobase interactions permits CcpA-(HPr-Ser46-P) to bind diverse operators. Indeed, biochemical data show that CcpA-(HPr-Ser46-P) binds the three cre sites with similar affinities. Thus, the data reveal properties that license this protein to function as a global transcription regulator.
Figures

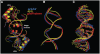


Similar articles
-
Phosphoprotein Crh-Ser46-P displays altered binding to CcpA to effect carbon catabolite regulation.J Biol Chem. 2006 Mar 10;281(10):6793-800. doi: 10.1074/jbc.M509977200. Epub 2005 Nov 29. J Biol Chem. 2006. PMID: 16316990
-
Structural basis for allosteric control of the transcription regulator CcpA by the phosphoprotein HPr-Ser46-P.Cell. 2004 Sep 17;118(6):731-41. doi: 10.1016/j.cell.2004.08.027. Cell. 2004. PMID: 15369672
-
Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors glucose 6-phosphate and fructose 1,6-bisphosphate.J Mol Biol. 2007 May 11;368(4):1042-50. doi: 10.1016/j.jmb.2007.02.054. Epub 2007 Feb 27. J Mol Biol. 2007. PMID: 17376479
-
Carbon catabolite control of the metabolic network in Bacillus subtilis.Biosci Biotechnol Biochem. 2009 Feb;73(2):245-59. doi: 10.1271/bbb.80479. Epub 2009 Feb 7. Biosci Biotechnol Biochem. 2009. PMID: 19202299 Review.
-
HPr kinase/phosphorylase, a Walker motif A-containing bifunctional sensor enzyme controlling catabolite repression in Gram-positive bacteria.Biochim Biophys Acta. 2004 Mar 11;1697(1-2):123-35. doi: 10.1016/j.bbapap.2003.11.018. Biochim Biophys Acta. 2004. PMID: 15023355 Review.
Cited by
-
Adaption to glucose limitation is modulated by the pleotropic regulator CcpA, independent of selection pressure strength.BMC Evol Biol. 2019 Jan 10;19(1):15. doi: 10.1186/s12862-018-1331-x. BMC Evol Biol. 2019. PMID: 30630406 Free PMC article.
-
CcpA represses the expression of the divergent cit operons of Enterococcus faecalis through multiple cre sites.BMC Microbiol. 2011 Oct 11;11:227. doi: 10.1186/1471-2180-11-227. BMC Microbiol. 2011. PMID: 21989394 Free PMC article.
-
A Novel Dual-cre Motif Enables Two-Way Autoregulation of CcpA in Clostridium acetobutylicum.Appl Environ Microbiol. 2018 Apr 2;84(8):e00114-18. doi: 10.1128/AEM.00114-18. Print 2018 Apr 15. Appl Environ Microbiol. 2018. PMID: 29427432 Free PMC article.
-
Probing the regulatory effects of specific mutations in three major binding domains of the pleiotropic regulator CcpA of Bacillus subtilis.Front Microbiol. 2015 Oct 2;6:1051. doi: 10.3389/fmicb.2015.01051. eCollection 2015. Front Microbiol. 2015. PMID: 26483775 Free PMC article.
-
Genome-wide analysis of in vivo CcpA binding with and without its key co-factor HPr in the major human pathogen group A Streptococcus.Mol Microbiol. 2021 Jun;115(6):1207-1228. doi: 10.1111/mmi.14667. Epub 2020 Dec 29. Mol Microbiol. 2021. PMID: 33325565 Free PMC article.
References
-
- Bruckner R, Titgemeyer F. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 2002;209:141–148. - PubMed
-
- Stülke J, Hillen W. Coupling physiology and gene regulation in bacteria: the phosphotransferase sugar uptake system delivers the signals. Naturwissenschaften. 1998;85:583–592. - PubMed
-
- Stülke J, Hillen W. Regulation of carbon catabolism in Bacillus species. Annu. Rev. Microbiol. 2000;54:849–880. - PubMed
-
- Fujita Y. Carbon catabolite control and the metabolic network in Bacillus subtilis. Biosci. Biotechnol. Biochem. 2009;73:245–259. - PubMed
-
- Deutscher J. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2008;11:87–93. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

