EUROCarbDB: An open-access platform for glycoinformatics
- PMID: 21106561
- PMCID: PMC3055595
- DOI: 10.1093/glycob/cwq188
EUROCarbDB: An open-access platform for glycoinformatics
Abstract
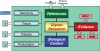
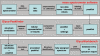
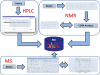
The EUROCarbDB project is a design study for a technical framework, which provides sophisticated, freely accessible, open-source informatics tools and databases to support glycobiology and glycomic research. EUROCarbDB is a relational database containing glycan structures, their biological context and, when available, primary and interpreted analytical data from high-performance liquid chromatography, mass spectrometry and nuclear magnetic resonance experiments. Database content can be accessed via a web-based user interface. The database is complemented by a suite of glycoinformatics tools, specifically designed to assist the elucidation and submission of glycan structure and experimental data when used in conjunction with contemporary carbohydrate research workflows. All software tools and source code are licensed under the terms of the Lesser General Public License, and publicly contributed structures and data are freely accessible. The public test version of the web interface to the EUROCarbDB can be found at http://www.ebi.ac.uk/eurocarb.
Figures


Similar articles
-
EUROCarbDB(CCRC): a EUROCarbDB node for storing glycomics standard data.Bioinformatics. 2015 Jan 15;31(2):242-5. doi: 10.1093/bioinformatics/btu609. Epub 2014 Sep 12. Bioinformatics. 2015. PMID: 25217575 Free PMC article.
-
Bioinformatics and molecular modeling in glycobiology.Cell Mol Life Sci. 2010 Aug;67(16):2749-72. doi: 10.1007/s00018-010-0352-4. Epub 2010 Apr 4. Cell Mol Life Sci. 2010. PMID: 20364395 Free PMC article. Review.
-
The Glycome Analytics Platform: an integrative framework for glycobioinformatics.Bioinformatics. 2016 Oct 1;32(19):3005-11. doi: 10.1093/bioinformatics/btw341. Epub 2016 Jun 10. Bioinformatics. 2016. PMID: 27288496
-
glypy: An Open Source Glycoinformatics Library.J Proteome Res. 2019 Sep 6;18(9):3532-3537. doi: 10.1021/acs.jproteome.9b00367. Epub 2019 Jul 30. J Proteome Res. 2019. PMID: 31310539 Free PMC article.
-
Glycoinformatics: Bridging Isolated Islands in the Sea of Data.Angew Chem Int Ed Engl. 2018 Nov 12;57(46):14986-14990. doi: 10.1002/anie.201803576. Epub 2018 Oct 11. Angew Chem Int Ed Engl. 2018. PMID: 29786940 Review.
Cited by
-
Comparison of Three Glycoproteomic Methods for the Analysis of the Secretome of CHO Cells Treated with 1,3,4-O-Bu3ManNAc.Bioengineering (Basel). 2020 Nov 10;7(4):144. doi: 10.3390/bioengineering7040144. Bioengineering (Basel). 2020. PMID: 33182731 Free PMC article.
-
The glycoconjugate ontology (GlycoCoO) for standardizing the annotation of glycoconjugate data and its application.Glycobiology. 2021 Aug 7;31(7):741-750. doi: 10.1093/glycob/cwab013. Glycobiology. 2021. PMID: 33677548 Free PMC article.
-
Property Graph vs RDF Triple Store: A Comparison on Glycan Substructure Search.PLoS One. 2015 Dec 14;10(12):e0144578. doi: 10.1371/journal.pone.0144578. eCollection 2015. PLoS One. 2015. PMID: 26656740 Free PMC article.
-
Protein Bioinformatics Databases and Resources.Methods Mol Biol. 2017;1558:3-39. doi: 10.1007/978-1-4939-6783-4_1. Methods Mol Biol. 2017. PMID: 28150231 Free PMC article. Review.
-
Strategies for the profiling, characterisation and detailed structural analysis of N-linked oligosaccharides.Glycoconj J. 2013 Feb;30(2):137-46. doi: 10.1007/s10719-012-9443-9. Epub 2012 Aug 26. Glycoconj J. 2013. PMID: 22922975 Review.
References
-
- Abd Hamid UM, Royle L, Saldova R, Radcliffe CM, Harvey DJ, Storr SJ, Pardo M, Antrobus R, Chapman CJ, Zitzmann N, et al. A strategy to reveal potential glycan markers from serum glycoproteins associated with breast cancer progression. Glycobiology. 2008;18:1105–1118. doi:10.1093/glycob/cwn095. - DOI - PubMed
-
- An HJ, Kronewitter SR, de Leoz ML, Lebrilla CB. Glycomics and disease markers. Curr Opin Chem Biol. 2009;13:601–607. doi:10.1016/j.cbpa.2009.08.015. - DOI - PMC - PubMed
-
- Aoki-Kinoshita KF. An introduction to bioinformatics for glycomics research. PLoS Comput Biol. 2008;4:e1000075. doi:10.1371/journal.pcbi.1000075. - DOI - PMC - PubMed
-
- Aoki KF, Yamaguchi A, Ueda N, Akutsu T, Mamitsuka H, Goto S, Kanehisa M. KCaM (KEGG Carbohydrate Matcher): A software tool for analyzing the structures of carbohydrate sugar chains. Nucleic Acids Res. 2004;32:W267–W272. doi:10.1093/nar/gkh473. - DOI - PMC - PubMed
-
- Arnold JN, Saldova R, Hamid UM, Rudd PM. Evaluation of the serum N-linked glycome for the diagnosis of cancer and chronic inflammation. Proteomics. 2008;8:3284–3293. doi:10.1002/pmic.200800163. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/F008309/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- B20259/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBF0083091/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/C519670/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/H004130/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom

