Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors
- PMID: 21106758
- PMCID: PMC3003021
- DOI: 10.1073/pnas.1013133107
Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors
Abstract
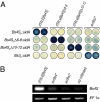
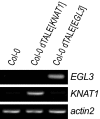
Proteins that can be tailored to bind desired DNA sequences are key tools for molecular biology. Previous studies suggested that DNA-binding specificity of transcription activator-like effectors (TALEs) from the bacterial genus Xanthomonas is defined by repeat-variable diresidues (RVDs) of tandem-arranged 34/35-amino acid repeat units. We have studied chimeras of two TALEs differing in RVDs and non-RVDs and found that, in contrast to the critical contributions by RVDs, non-RVDs had no major effect on the DNA-binding specificity of the chimeras. This finding suggests that one needs only to modify the RVDs to generate designer TALEs (dTALEs) to activate transcription of user-defined target genes. We used the scaffold of the TALE AvrBs3 and changed its RVDs to match either the tomato Bs4, the Arabidopsis EGL3, or the Arabidopsis KNAT1 promoter. All three dTALEs transcriptionally activated the desired promoters in a sequence-specific manner as mutations within the targeted DNA sequences abolished promoter activation. This study is unique in showing that chromosomal loci can be targeted specifically by dTALEs. We also engineered two AvrBs3 derivatives with four additional repeat units activating specifically either the pepper Bs3 or UPA20 promoter. Because AvrBs3 activates both promoters, our data show that addition of repeat units facilitates TALE-specificity fine-tuning. Finally, we demonstrate that the RVD NK mediates specific interaction with G nucleotides that thus far could not be targeted specifically by any known RVD type. In summary, our data demonstrate that the TALE scaffold can be tailored to target user-defined DNA sequences in whole genomes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. - PubMed
-
- Kay S, Hahn S, Marois E, Hause G, Bonas U. A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science. 2007;318:648–651. - PubMed
-
- Römer P, et al. Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science. 2007;318:645–648. - PubMed
-
- Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol. 2010;48:419–436. - PubMed
-
- Bogdanove AJ, Schornack S, Lahaye T. TAL effectors: Finding plant genes for disease and defense. Curr Opin Plant Biol. 2010;13:394–401. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

