Multilocus sequence analysis and rpoB sequencing of Mycobacterium abscessus (sensu lato) strains
- PMID: 21106786
- PMCID: PMC3043527
- DOI: 10.1128/JCM.01274-10
Multilocus sequence analysis and rpoB sequencing of Mycobacterium abscessus (sensu lato) strains
Abstract
Mycobacterium abscessus, Mycobacterium bolletii, and Mycobacterium massiliense (Mycobacterium abscessus sensu lato) are closely related species that currently are identified by the sequencing of the rpoB gene. However, recent studies show that rpoB sequencing alone is insufficient to discriminate between these species, and some authors have questioned their current taxonomic classification. We studied here a large collection of M. abscessus (sensu lato) strains by partial rpoB sequencing (752 bp) and multilocus sequence analysis (MLSA). The final MLSA scheme developed was based on the partial sequences of eight housekeeping genes: argH, cya, glpK, gnd, murC, pgm, pta, and purH. The strains studied included the three type strains (M. abscessus CIP 104536(T), M. massiliense CIP 108297(T), and M. bolletii CIP 108541(T)) and 120 isolates recovered between 1997 and 2007 in France, Germany, Switzerland, and Brazil. The rpoB phylogenetic tree confirmed the existence of three main clusters, each comprising the type strain of one species. However, divergence values between the M. massiliense and M. bolletii clusters all were below 3% and between the M. abscessus and M. massiliense clusters were from 2.66 to 3.59%. The tree produced using the concatenated MLSA gene sequences (4,071 bp) also showed three main clusters, each comprising the type strain of one species. The M. abscessus cluster had a bootstrap value of 100% and was mostly compact. Bootstrap values for the M. massiliense and M. bolletii branches were much lower (71 and 61%, respectively), with the M. massiliense cluster having a fuzzy aspect. Mean (range) divergence values were 2.17% (1.13 to 2.58%) between the M. abscessus and M. massiliense clusters, 2.37% (1.5 to 2.85%) between the M. abscessus and M. bolletii clusters, and 2.28% (0.86 to 2.68%) between the M. massiliense and M. bolletii clusters. Adding the rpoB sequence to the MLSA-concatenated sequence (total sequence, 4,823 bp) had little effect on the clustering of strains. We found 10/120 (8.3%) isolates for which the concatenated MLSA gene sequence and rpoB sequence were discordant (e.g., M. massiliense MLSA sequence and M. abscessus rpoB sequence), suggesting the intergroup lateral transfers of rpoB. In conclusion, our study strongly supports the recent proposal that M. abscessus, M. massiliense, and M. bolletii should constitute a single species. Our findings also indicate that there has been a horizontal transfer of rpoB sequences between these subgroups, precluding the use of rpoB sequencing alone for the accurate identification of the two proposed M. abscessus subspecies.
Figures
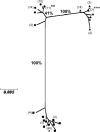
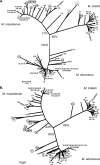
References
-
- Adekambi T., Berger P., Raoult D., Drancourt M. 2006. rpoB gene sequence-based characterization of emerging nontuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov. Int. J. Syst. Evol. Microbiol. 56:133–143 - PubMed
-
- Adékambi T., Drancourt M. 2004. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA and rpoB gene sequencing. Int. J. Syst. Evol. Microbiol. 54:2095–2105 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

