High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells
- PMID: 21106903
- PMCID: PMC3044859
- DOI: 10.1101/gr.112656.110
High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells
Abstract
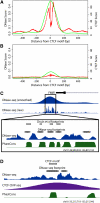
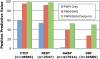
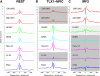
Regulation of gene transcription in diverse cell types is determined largely by varied sets of cis-elements where transcription factors bind. Here we demonstrate that data from a single high-throughput DNase I hypersensitivity assay can delineate hundreds of thousands of base-pair resolution in vivo footprints in human cells that precisely mark individual transcription factor-DNA interactions. These annotations provide a unique resource for the investigation of cis-regulatory elements. We find that footprints for specific transcription factors correlate with ChIP-seq enrichment and can accurately identify functional versus nonfunctional transcription factor motifs. We also find that footprints reveal a unique evolutionary conservation pattern that differentiates functional footprinted bases from surrounding DNA. Finally, detailed analysis of CTCF footprints suggests multiple modes of binding and a novel DNA binding motif upstream of the primary binding site.
Figures
References
-
- Boffelli D, McAuliffe J, Ovcharenko D, Lewis KD, Ovcharenko I, Pachter L, Rubin EM 2003. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299: 1391–1394 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
