Tension directly stabilizes reconstituted kinetochore-microtubule attachments
- PMID: 21107429
- PMCID: PMC3108429
- DOI: 10.1038/nature09594
Tension directly stabilizes reconstituted kinetochore-microtubule attachments
Abstract
Kinetochores are macromolecular machines that couple chromosomes to dynamic microtubule tips during cell division, thereby generating force to segregate the chromosomes. Accurate segregation depends on selective stabilization of correct 'bi-oriented' kinetochore-microtubule attachments, which come under tension as the result of opposing forces exerted by microtubules. Tension is thought to stabilize these bi-oriented attachments indirectly, by suppressing the destabilizing activity of a kinase, Aurora B. However, a complete mechanistic understanding of the role of tension requires reconstitution of kinetochore-microtubule attachments for biochemical and biophysical analyses in vitro. Here we show that native kinetochore particles retaining the majority of kinetochore proteins can be purified from budding yeast and used to reconstitute dynamic microtubule attachments. Individual kinetochore particles maintain load-bearing associations with assembling and disassembling ends of single microtubules for >30 min, providing a close match to the persistent coupling seen in vivo between budding yeast kinetochores and single microtubules. Moreover, tension increases the lifetimes of the reconstituted attachments directly, through a catch bond-like mechanism that does not require Aurora B. On the basis of these findings, we propose that tension selectively stabilizes proper kinetochore-microtubule attachments in vivo through a combination of direct mechanical stabilization and tension-dependent phosphoregulation.
Figures


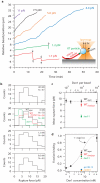
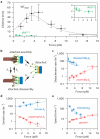
Comment in
-
Mechanoregulation: Cellular seat belts.Nature. 2010 Nov 25;468(7323):518-9. doi: 10.1038/468518a. Nature. 2010. PMID: 21107420 No abstract available.
References
Methods Additional References
-
- Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA015704/CA/NCI NIH HHS/United States
- T32HL007312/HL/NHLBI NIH HHS/United States
- GM078069/GM/NIGMS NIH HHS/United States
- T32 HL007312/HL/NHLBI NIH HHS/United States
- R01 GM079373/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- T32 GM007270/GM/NIGMS NIH HHS/United States
- P50 GM076547/GM/NIGMS NIH HHS/United States
- GM064386/GM/NIGMS NIH HHS/United States
- T32GM07270/GM/NIGMS NIH HHS/United States
- PM50 GM076547/GM/NIGMS NIH HHS/United States
- R01GM79373/GM/NIGMS NIH HHS/United States
- P30 CA015704/CA/NCI NIH HHS/United States
- R01 GM078069/GM/NIGMS NIH HHS/United States
- R01 GM064386/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

