Intronic miR-211 assumes the tumor suppressive function of its host gene in melanoma
- PMID: 21109473
- PMCID: PMC3004467
- DOI: 10.1016/j.molcel.2010.11.020
Intronic miR-211 assumes the tumor suppressive function of its host gene in melanoma
Abstract
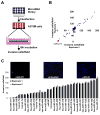
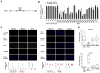
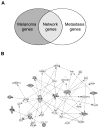
When it escapes early detection, malignant melanoma becomes a highly lethal and treatment-refractory cancer. Melastatin is greatly downregulated in metastatic melanomas and is widely believed to function as a melanoma tumor suppressor. Here we report that tumor suppressive activity is not mediated by melastatin but instead by a microRNA (miR-211) hosted within an intron of melastatin. Increasing expression of miR-211 but not melastatin reduced migration and invasion of malignant and highly invasive human melanomas characterized by low levels of melastatin and miR-211. An unbiased network analysis of melanoma-expressed genes filtered for their roles in metastasis identified three central node genes: IGF2R, TGFBR2, and NFAT5. Expression of these genes was reduced by miR-211, and knockdown of each gene phenocopied the effects of increased miR-211 on melanoma invasiveness. These data implicate miR-211 as a suppressor of melanoma invasion whose expression is silenced or selected against via suppression of the entire melastatin locus during human melanoma progression.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures

References
-
- Deeds J, Cronin F, Duncan LM. Patterns of melastatin mRNA expression in melanocytic tumors. Hum Pathol. 2000;31:1346–1356. - PubMed
-
- Duncan LM, Deeds J, Cronin FE, Donovan M, Sober AJ, Kauffman M, McCarthy JJ. Melastatin expression and prognosis in cutaneous malignant melanoma. J Clin Oncol. 2001;19:568–576. - PubMed
-
- Duncan LM, Deeds J, Hunter J, Shao J, Holmgren LM, Woolf EA, Tepper RI, Shyjan AW. Down-regulation of the novel gene melastatin correlates with potential for melanoma metastasis. Cancer Res. 1998;58:1515–1520. - PubMed
-
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. - PubMed
-
- Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, Ambros VR, Israel MA. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67:2456–2468. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

