Endogenous ribosomal frameshift signals operate as mRNA destabilizing elements through at least two molecular pathways in yeast
- PMID: 21109528
- PMCID: PMC3074144
- DOI: 10.1093/nar/gkq1220
Endogenous ribosomal frameshift signals operate as mRNA destabilizing elements through at least two molecular pathways in yeast
Abstract
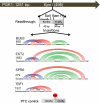
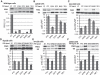
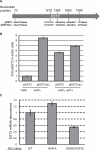
Although first discovered in viruses, previous studies have identified operational -1 ribosomal frameshifting (-1 RF) signals in eukaryotic genomic sequences, and suggested a role in mRNA stability. Here, four yeast -1 RF signals are shown to promote significant mRNA destabilization through the nonsense mediated mRNA decay pathway (NMD), and genetic evidence is presented suggesting that they may also operate through the no-go decay pathway (NGD) as well. Yeast EST2 mRNA is highly unstable and contains up to five -1 RF signals. Ablation of the -1 RF signals or of NMD stabilizes this mRNA, and changes in -1 RF efficiency have opposing effects on the steady-state abundance of the EST2 mRNA. These results demonstrate that endogenous -1 RF signals function as mRNA destabilizing elements through at least two molecular pathways in yeast. Consistent with current evolutionary theory, phylogenetic analyses suggest that -1 RF signals are rapidly evolving cis-acting regulatory elements. Identification of high confidence -1 RF signals in ∼10% of genes in all eukaryotic genomes surveyed suggests that -1 RF is a broadly used post-transcriptional regulator of gene expression.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

