Plasmodium falciparum genetic diversity maintained and amplified over 5 years of a low transmission endemic in the Peruvian Amazon
- PMID: 21109587
- PMCID: PMC3112368
- DOI: 10.1093/molbev/msq311
Plasmodium falciparum genetic diversity maintained and amplified over 5 years of a low transmission endemic in the Peruvian Amazon
Abstract
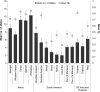
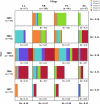
Plasmodium falciparum entered into the Peruvian Amazon in 1994, sparking an epidemic between 1995 and 1998. Since 2000, there has been sustained low P. falciparum transmission. The Malaria Immunology and Genetics in the Amazon project has longitudinally followed members of the community of Zungarococha (N = 1,945, 4 villages) with active household and health center-based visits each year since 2003. We examined parasite population structure and traced the parasite genetic diversity temporally and spatially. We genotyped infections over 5 years (2003-2007) using 14 microsatellite (MS) markers scattered across ten different chromosomes. Despite low transmission, there was considerable genetic diversity, which we compared with other geographic regions. We detected 182 different haplotypes from 302 parasites in 217 infections. Structure v2.2 identified five clusters (subpopulations) of phylogenetically related clones. To consider genetic diversity on a more detailed level, we defined haplotype families (hapfams) by grouping haplotypes with three or less loci differences. We identified 34 different hapfams identified. The F(st) statistic and heterozygosity analysis showed the five clusters were maintained in each village throughout this time. A minimum spanning network (MSN), stratified by the year of detection, showed that haplotypes within hapfams had allele differences and haplotypes within a cluster definition were more separated in the later years (2006-2007). We modeled hapfam detection and loss, accounting for sample size and stochastic fluctuations in frequencies overtime. Principle component analysis of genetic variation revealed patterns of genetic structure with time rather than village. The population structure, genetic diversity, appearance/disappearance of the different haplotypes from 2003 to 2007 provides a genome-wide "real-time" perspective of P. falciparum parasites in a low transmission region.
Figures



References
-
- Anderson TJ, Haubold B, Williams JT, et al. (16 co-authors) Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000;17:1467–1482. - PubMed
-
- Bogreau H, Renaud F, Bouchiba H, et al. (15 co-authors) Genetic diversity and structure of African Plasmodium falciparum populations in urban and rural areas. Am J Trop Med Hyg. 2006;74:953–959. - PubMed
-
- Branch O, Casapia WM, Gamboa DV, Hernandez JN, Alava FF, Roncal N, Alvarez E, Perez EJ, Gotuzzo E. Clustered local transmission and asymptomatic Plasmodium falciparum and Plasmodium vivax malaria infections in a recently emerged, hypoendemic Peruvian Amazon community. Malar J. 2005;4:27. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

