Mechanisms determining the morphology of the peripheral ER
- PMID: 21111237
- PMCID: PMC3008339
- DOI: 10.1016/j.cell.2010.11.007
Mechanisms determining the morphology of the peripheral ER
Abstract
The endoplasmic reticulum (ER) consists of the nuclear envelope and a peripheral network of tubules and membrane sheets. The tubules are shaped by the curvature-stabilizing proteins reticulons and DP1/Yop1p, but how the sheets are formed is unclear. Here, we identify several sheet-enriched membrane proteins in the mammalian ER, including proteins that translocate and modify newly synthesized polypeptides, as well as coiled-coil membrane proteins that are highly upregulated in cells with proliferated ER sheets, all of which are localized by membrane-bound polysomes. These results indicate that sheets and tubules correspond to rough and smooth ER, respectively. One of the coiled-coil proteins, Climp63, serves as a "luminal ER spacer" and forms sheets when overexpressed. More universally, however, sheet formation appears to involve the reticulons and DP1/Yop1p, which localize to sheet edges and whose abundance determines the ratio of sheets to tubules. These proteins may generate sheets by stabilizing the high curvature of edges.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures

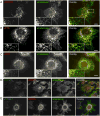
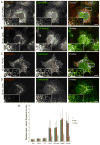



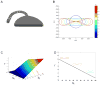
Comment in
-
ER sheets get roughed up.Cell. 2010 Nov 24;143(5):665-6. doi: 10.1016/j.cell.2010.11.011. Cell. 2010. PMID: 21111224
References
-
- Ambroziak J, Henry SA. INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J Biol Chem. 1994;269:15344–15349. - PubMed
-
- Baumann O, Walz B. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int Rev Cytol. 2001;205:149–214. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

