Metagenomics and the molecular identification of novel viruses
- PMID: 21111643
- PMCID: PMC7110547
- DOI: 10.1016/j.tvjl.2010.10.014
Metagenomics and the molecular identification of novel viruses
Abstract
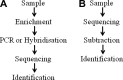
There have been rapid recent developments in establishing methods for identifying and characterising viruses associated with animal and human diseases. These methodologies, commonly based on hybridisation or PCR techniques, are combined with advanced sequencing techniques termed 'next generation sequencing'. Allied advances in data analysis, including the use of computational transcriptome subtraction, have also impacted the field of viral pathogen discovery. This review details these molecular detection techniques, discusses their application in viral discovery, and provides an overview of some of the novel viruses discovered. The problems encountered in attributing disease causality to a newly identified virus are also considered.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures


Comment in
-
Pushing the envelope: advances in molecular techniques for the detection of novel viruses.Vet J. 2011 Nov;190(2):185-186. doi: 10.1016/j.tvjl.2011.02.002. Epub 2011 Mar 10. Vet J. 2011. PMID: 21396861 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

