Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips
- PMID: 21113165
- PMCID: PMC3139991
- DOI: 10.1038/nbt.1716
Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips
Abstract
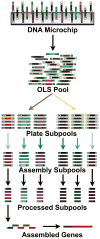
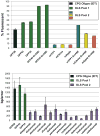
Development of cheap, high-throughput and reliable gene synthesis methods will broadly stimulate progress in biology and biotechnology. Currently, the reliance on column-synthesized oligonucleotides as a source of DNA limits further cost reductions in gene synthesis. Oligonucleotides from DNA microchips can reduce costs by at least an order of magnitude, yet efforts to scale their use have been largely unsuccessful owing to the high error rates and complexity of the oligonucleotide mixtures. Here we use high-fidelity DNA microchips, selective oligonucleotide pool amplification, optimized gene assembly protocols and enzymatic error correction to develop a method for highly parallel gene synthesis. We tested our approach by assembling 47 genes, including 42 challenging therapeutic antibody sequences, encoding a total of ∼35 kilobase pairs of DNA. These assemblies were performed from a complex background containing 13,000 oligonucleotides encoding ∼2.5 megabases of DNA, which is at least 50 times larger than in previously published attempts.
Conflict of interest statement
E.M.L. is an employee of Agilent Technologies, the commercial provider of OLS pools. G.M.C. is a co-founder of an early-stage startup company involved in gene synthesis. S.K., N.E., and G.M.C. are named inventors on a patent application on technologies described in this article. S.K. is a post-doctoral fellow whose future employment prospects depend upon refereed publications.
Figures

Comment in
-
Making more genes, better and for less.Nat Rev Genet. 2011 Jan;12(1):6. doi: 10.1038/nrg2924. Nat Rev Genet. 2011. PMID: 21164523 No abstract available.
References
-
- Carr PA, Church GM. Genome engineering. Nat Biotechnol. 2009;27:1151–1162. - PubMed
-
- Tian J, Ma K, Saaem I. Advancing high-throughput gene synthesis technology. Mol BioSyst. 2009;5:714–722. - PubMed
-
- Tian J, et al. Accurate multiplex gene synthesis from programmable DNA microchips. Nature. 2004;432:1050–1054. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

