The histone methyltransferase MLL1 permits the oscillation of circadian gene expression
- PMID: 21113167
- PMCID: PMC6501791
- DOI: 10.1038/nsmb.1961
The histone methyltransferase MLL1 permits the oscillation of circadian gene expression
Abstract
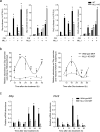
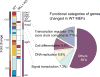
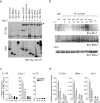
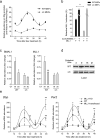
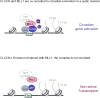
The classical view of the molecular clock is based on interlocked transcriptional-translational feedback loops. Because a substantial fraction of the mammalian genome is expressed in a circadian manner, chromatin remodeling has been proposed to be crucial in clock function. Here we show that Lys4 (K4) trimethylation of histone H3 is rhythmic and follows the same profile as previously described H3 acetylation on circadian promoters. MLL1, a mammalian homolog of Drosophila trithorax, is an H3K4-specific methyltransferase implicated in transcriptional control. We demonstrate that MLL1 is essential for circadian transcription and cyclic H3K4 trimethylation. MLL1 is in a complex with CLOCK-BMAL1 and contributes to its rhythmic recruitment to circadian promoters and to H3 acetylation. Yet MLL1 fails to interact with CLOCKΔ19, providing an explanation for this mutation's dominant negative phenotype. Our results favor a scenario in which H3K4 trimethylation by MLL1 is required to establish a permissive chromatin state for circadian transcription.
Figures

References
-
- Dunlap JC, Loros JJ, Liu Y, Crosthwaite SK. Eukaryotic circadian systems: cycles in common. Genes Cells. 1999;4:1–10. - PubMed
-
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–41. - PubMed
-
- Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–22. - PubMed
-
- Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3:350–61. - PubMed

