Relative free energy of binding between antimicrobial peptides and SDS or DPC micelles
- PMID: 21113423
- PMCID: PMC2990536
- DOI: 10.1080/08927020902902742
Relative free energy of binding between antimicrobial peptides and SDS or DPC micelles
Abstract
We present relative binding free energy calculations for six antimicrobial peptide-micelle systems, three peptides interacting with two types of micelles. The peptides are the scorpion derived antimicrobial peptide (AMP), IsCT and two of its analogues. The micelles are dodecylphosphatidylcholine (DPC) and sodium dodecylsulphate (SDS) micelles. The interfacial electrostatic properties of DPC and SDS micelles are assumed to be similar to those of zwitterionic mammalian and anionic bacterial membrane interfaces, respectively. We test the hypothesis that the binding strength between peptides and the anionic micelle SDS can provide information on peptide antimicrobial activity, since it is widely accepted that AMPs function by binding to and disrupting the predominantly anionic lipid bilayer of the bacterial cytoplasmic membrane. We also test the hypothesis that the binding strength between peptides and the zwitterionic micelle DPC can provide information on peptide haemolytic activities, since it is accepted that they also bind to and disrupt the zwitterionic membrane of mammalian cells. Equilibrium structures of the peptides, micelles and peptide-micelle complexes are obtained from more than 300 ns of molecular dynamics simulations. A thermodynamic cycle is introduced to compute the binding free energy from electrostatic, non-electrostatic and entropic contributions. We find relative binding free energy strengths between peptides and SDS to correlate with the experimentally measured rankings for peptide antimicrobial activities, and relative free energy binding strengths between peptides and DPC to correlate with the observed rankings for peptide haemolytic toxicities. These findings point to the importance of peptide-membrane binding strength for antimicrobial activity and haemolytic activity.
Figures






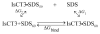
Similar articles
-
Effect of Leu/Val Mutation on the Energetics of Antimicrobial Peptide:Micelle Binding.J Phys Chem B. 2022 Jul 21;126(28):5262-5273. doi: 10.1021/acs.jpcb.2c01293. Epub 2022 Jul 10. J Phys Chem B. 2022. PMID: 35815580
-
Molecular dynamics investigation of the influence of anionic and zwitterionic interfaces on antimicrobial peptides' structure: implications for peptide toxicity and activity.Peptides. 2006 Jun;27(6):1192-200. doi: 10.1016/j.peptides.2005.10.022. Epub 2005 Dec 1. Peptides. 2006. PMID: 16325306 Free PMC article.
-
Antimicrobial peptide RP-1 structure and interactions with anionic versus zwitterionic micelles.Biopolymers. 2009 Jan;91(1):1-13. doi: 10.1002/bip.21071. Biopolymers. 2009. PMID: 18712851
-
Structural characterization of the antimicrobial peptides myxinidin and WMR in bacterial membrane mimetic micelles and bicelles.Biochim Biophys Acta Biomembr. 2024 Mar;1866(3):184272. doi: 10.1016/j.bbamem.2024.184272. Epub 2024 Jan 9. Biochim Biophys Acta Biomembr. 2024. PMID: 38211645 Review.
-
Solution NMR studies of amphibian antimicrobial peptides: linking structure to function?Biochim Biophys Acta. 2009 Aug;1788(8):1639-55. doi: 10.1016/j.bbamem.2009.01.002. Epub 2009 Jan 15. Biochim Biophys Acta. 2009. PMID: 19272309 Review.
Cited by
-
A Critical Review of Short Antimicrobial Peptides from Scorpion Venoms, Their Physicochemical Attributes, and Potential for the Development of New Drugs.J Membr Biol. 2024 Aug;257(3-4):165-205. doi: 10.1007/s00232-024-00315-2. Epub 2024 Jul 11. J Membr Biol. 2024. PMID: 38990274 Free PMC article. Review.
-
Perturbations of Native Membrane Protein Structure in Alkyl Phosphocholine Detergents: A Critical Assessment of NMR and Biophysical Studies.Chem Rev. 2018 Apr 11;118(7):3559-3607. doi: 10.1021/acs.chemrev.7b00570. Epub 2018 Feb 28. Chem Rev. 2018. PMID: 29488756 Free PMC article. Review.
-
Molecular and energetic analysis of the interaction and specificity of Maximin 3 with lipid membranes: In vitro and in silico assessments.Protein Sci. 2024 Nov;33(11):e5188. doi: 10.1002/pro.5188. Protein Sci. 2024. PMID: 39473071
-
Multiscale models of the antimicrobial peptide protegrin-1 on gram-negative bacteria membranes.Int J Mol Sci. 2012;13(9):11000-11011. doi: 10.3390/ijms130911000. Epub 2012 Sep 5. Int J Mol Sci. 2012. PMID: 23109834 Free PMC article. Review.
-
Designing Antibacterial Peptides with Enhanced Killing Kinetics.Front Microbiol. 2018 Feb 23;9:325. doi: 10.3389/fmicb.2018.00325. eCollection 2018. Front Microbiol. 2018. PMID: 29527201 Free PMC article.
References
-
- Yoshikawa TT. Antimicrobial resistance and aging: beginning of the end of the antibiotic era? J Am Geriatr Soc. 2002;50:226–229. - PubMed
-
- Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover FC. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997;40:135–136. - PubMed
-
- Pallares R, Linares J, Vadillo M, Cabellos C, Manresa F, Viladrich PF, Martin R, Gudiol F. Resistance to penicillin and cephalosporin and mortality from severe pneumococcal pneumonia in Barcelona, Spain. N Engl J Med. 1995;333:474–480. - PubMed
-
- Grubb W. Genetics of MRSA. Rev Med Microbiol. 1998;9:153–162.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
